
Oscillibacter sp. CAG:241
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Oscillibacter; environmental samples
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
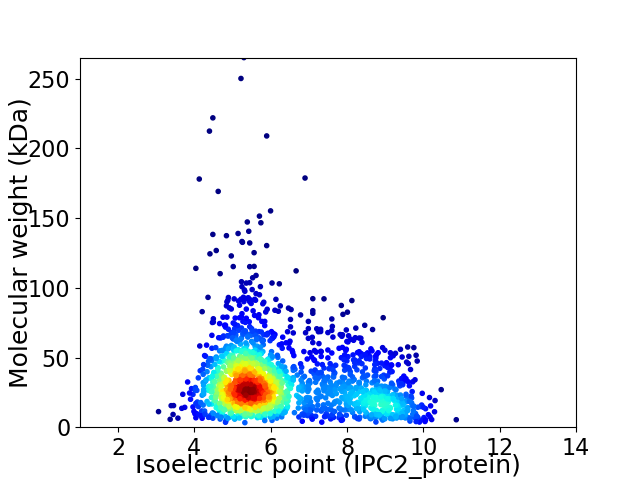
Virtual 2D-PAGE plot for 1823 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
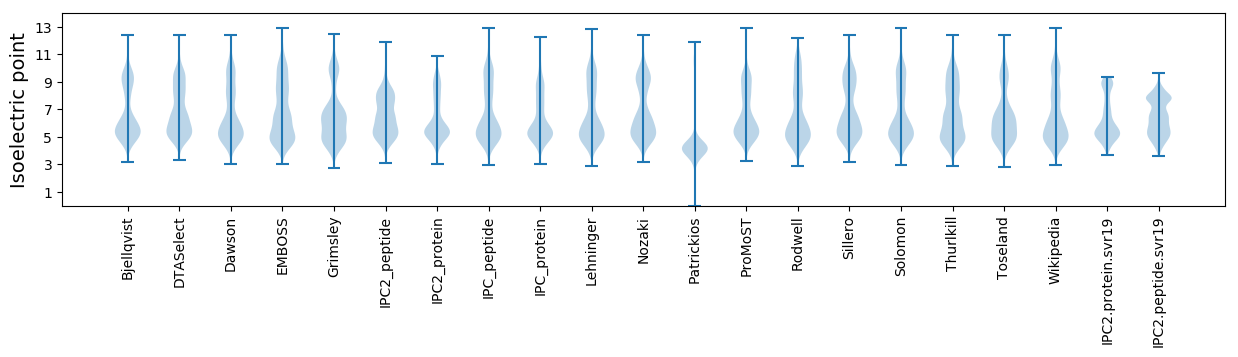
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6HG37|R6HG37_9FIRM Histidine kinase OS=Oscillibacter sp. CAG:241 OX=1262911 GN=BN557_01341 PE=4 SV=1
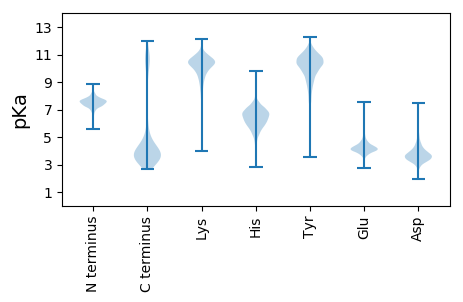
MM1 pKa = 7.53KK2 pKa = 10.22KK3 pKa = 10.56LSRR6 pKa = 11.84LAVMLALVLALTACSGGADD25 pKa = 3.55TPSPDD30 pKa = 3.34EE31 pKa = 4.06QTEE34 pKa = 4.3GQHH37 pKa = 6.05PVTDD41 pKa = 3.71AAEE44 pKa = 5.12LEE46 pKa = 4.59GQWTSPSGNILSFSADD62 pKa = 3.09QGRR65 pKa = 11.84YY66 pKa = 6.99TYY68 pKa = 9.21QTFSGRR74 pKa = 11.84TGQGPYY80 pKa = 10.2DD81 pKa = 3.7AAADD85 pKa = 3.8VPTIDD90 pKa = 4.97FDD92 pKa = 3.78GFLYY96 pKa = 10.75DD97 pKa = 6.16FILRR101 pKa = 11.84DD102 pKa = 4.57DD103 pKa = 4.69GVLLPNQNGSGGDD116 pKa = 3.57AEE118 pKa = 5.32SIDD121 pKa = 3.47HH122 pKa = 5.16FTFRR126 pKa = 11.84RR127 pKa = 11.84SSDD130 pKa = 2.87ARR132 pKa = 11.84IDD134 pKa = 3.4LWEE137 pKa = 4.6LSDD140 pKa = 6.31LSGVWQNALGEE151 pKa = 4.39TLVIDD156 pKa = 4.9ADD158 pKa = 3.81RR159 pKa = 11.84MAYY162 pKa = 10.49LSYY165 pKa = 10.57TRR167 pKa = 11.84LSMGSGTIGDD177 pKa = 4.17DD178 pKa = 3.47QDD180 pKa = 4.02GKK182 pKa = 11.44GPYY185 pKa = 9.91LAMNGRR191 pKa = 11.84IYY193 pKa = 10.83LSPGLDD199 pKa = 3.06KK200 pKa = 11.42DD201 pKa = 3.75RR202 pKa = 11.84FTLLGQDD209 pKa = 3.3ADD211 pKa = 4.07GSFSGVFYY219 pKa = 10.04PYY221 pKa = 10.45GQAAAYY227 pKa = 6.76TALPGASFEE236 pKa = 4.22TDD238 pKa = 2.57EE239 pKa = 5.9DD240 pKa = 5.9GYY242 pKa = 10.99CRR244 pKa = 11.84WYY246 pKa = 11.14SDD248 pKa = 3.04GHH250 pKa = 5.89DD251 pKa = 3.28AFYY254 pKa = 9.58ITGSYY259 pKa = 8.89TLAEE263 pKa = 5.13DD264 pKa = 3.74GLAYY268 pKa = 10.34RR269 pKa = 11.84DD270 pKa = 4.4DD271 pKa = 4.59GPVYY275 pKa = 10.57AAGFDD280 pKa = 4.49PIPYY284 pKa = 10.11DD285 pKa = 3.92PAEE288 pKa = 4.46DD289 pKa = 3.91WGDD292 pKa = 2.87SWMSNWGG299 pKa = 3.36
MM1 pKa = 7.53KK2 pKa = 10.22KK3 pKa = 10.56LSRR6 pKa = 11.84LAVMLALVLALTACSGGADD25 pKa = 3.55TPSPDD30 pKa = 3.34EE31 pKa = 4.06QTEE34 pKa = 4.3GQHH37 pKa = 6.05PVTDD41 pKa = 3.71AAEE44 pKa = 5.12LEE46 pKa = 4.59GQWTSPSGNILSFSADD62 pKa = 3.09QGRR65 pKa = 11.84YY66 pKa = 6.99TYY68 pKa = 9.21QTFSGRR74 pKa = 11.84TGQGPYY80 pKa = 10.2DD81 pKa = 3.7AAADD85 pKa = 3.8VPTIDD90 pKa = 4.97FDD92 pKa = 3.78GFLYY96 pKa = 10.75DD97 pKa = 6.16FILRR101 pKa = 11.84DD102 pKa = 4.57DD103 pKa = 4.69GVLLPNQNGSGGDD116 pKa = 3.57AEE118 pKa = 5.32SIDD121 pKa = 3.47HH122 pKa = 5.16FTFRR126 pKa = 11.84RR127 pKa = 11.84SSDD130 pKa = 2.87ARR132 pKa = 11.84IDD134 pKa = 3.4LWEE137 pKa = 4.6LSDD140 pKa = 6.31LSGVWQNALGEE151 pKa = 4.39TLVIDD156 pKa = 4.9ADD158 pKa = 3.81RR159 pKa = 11.84MAYY162 pKa = 10.49LSYY165 pKa = 10.57TRR167 pKa = 11.84LSMGSGTIGDD177 pKa = 4.17DD178 pKa = 3.47QDD180 pKa = 4.02GKK182 pKa = 11.44GPYY185 pKa = 9.91LAMNGRR191 pKa = 11.84IYY193 pKa = 10.83LSPGLDD199 pKa = 3.06KK200 pKa = 11.42DD201 pKa = 3.75RR202 pKa = 11.84FTLLGQDD209 pKa = 3.3ADD211 pKa = 4.07GSFSGVFYY219 pKa = 10.04PYY221 pKa = 10.45GQAAAYY227 pKa = 6.76TALPGASFEE236 pKa = 4.22TDD238 pKa = 2.57EE239 pKa = 5.9DD240 pKa = 5.9GYY242 pKa = 10.99CRR244 pKa = 11.84WYY246 pKa = 11.14SDD248 pKa = 3.04GHH250 pKa = 5.89DD251 pKa = 3.28AFYY254 pKa = 9.58ITGSYY259 pKa = 8.89TLAEE263 pKa = 5.13DD264 pKa = 3.74GLAYY268 pKa = 10.34RR269 pKa = 11.84DD270 pKa = 4.4DD271 pKa = 4.59GPVYY275 pKa = 10.57AAGFDD280 pKa = 4.49PIPYY284 pKa = 10.11DD285 pKa = 3.92PAEE288 pKa = 4.46DD289 pKa = 3.91WGDD292 pKa = 2.87SWMSNWGG299 pKa = 3.36
Molecular weight: 32.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6GU42|R6GU42_9FIRM Uncharacterized protein OS=Oscillibacter sp. CAG:241 OX=1262911 GN=BN557_01473 PE=4 SV=1
MM1 pKa = 7.89DD2 pKa = 4.0KK3 pKa = 10.01WKK5 pKa = 9.21KK6 pKa = 9.18TIMHH10 pKa = 6.53MGAYY14 pKa = 8.22PAALGKK20 pKa = 9.24WVLLGGIIGVAGGVIGSLFHH40 pKa = 8.19IGVNYY45 pKa = 8.4ATAIRR50 pKa = 11.84GAHH53 pKa = 5.93PWILYY58 pKa = 7.62LLPVGGLAIVGLYY71 pKa = 9.65KK72 pKa = 10.54LCHH75 pKa = 6.72LEE77 pKa = 4.27GKK79 pKa = 8.16GTNAIIEE86 pKa = 4.44SVHH89 pKa = 6.84FGEE92 pKa = 5.05SVPILLVPVIFVSTVITHH110 pKa = 6.78LCGGSAGRR118 pKa = 11.84EE119 pKa = 3.93GAALQIGGGLGFQAGKK135 pKa = 10.35LLRR138 pKa = 11.84LGEE141 pKa = 4.34KK142 pKa = 10.13DD143 pKa = 4.13LPLATLCGMSGVFSALFGTPLTATVFALEE172 pKa = 4.38VISVGVLYY180 pKa = 10.84YY181 pKa = 10.89AGLVPCITAAMAAFGVSALMGVEE204 pKa = 3.98PTRR207 pKa = 11.84FTVSMPRR214 pKa = 11.84VTLEE218 pKa = 3.48LMLPVVVLSILCALVSILFCKK239 pKa = 9.44GLHH242 pKa = 4.59WTEE245 pKa = 4.12HH246 pKa = 5.85LLTRR250 pKa = 11.84SFRR253 pKa = 11.84NPWLRR258 pKa = 11.84VLAGAAILIVLSLLTNGDD276 pKa = 3.78YY277 pKa = 11.23NGAGMDD283 pKa = 3.86VIARR287 pKa = 11.84ALRR290 pKa = 11.84GNVSGWAWLGEE301 pKa = 4.21VLFFPLPPLLGAVHH315 pKa = 6.67RR316 pKa = 11.84RR317 pKa = 11.84HH318 pKa = 6.27HH319 pKa = 6.47RR320 pKa = 11.84LRR322 pKa = 11.84LQGRR326 pKa = 11.84RR327 pKa = 11.84GGAVVLRR334 pKa = 11.84GRR336 pKa = 11.84CLRR339 pKa = 11.84LLHH342 pKa = 6.62GGAPGPACGLCRR354 pKa = 11.84GHH356 pKa = 6.92RR357 pKa = 11.84PGGGVLRR364 pKa = 11.84GGEE367 pKa = 4.12LPRR370 pKa = 11.84GVGAAQRR377 pKa = 11.84GAVRR381 pKa = 11.84LCWGALLRR389 pKa = 11.84RR390 pKa = 11.84GLRR393 pKa = 11.84PEE395 pKa = 4.59LSPLRR400 pKa = 11.84LLRR403 pKa = 11.84PVQQPDD409 pKa = 3.25HH410 pKa = 7.12PLFQAPCGVHH420 pKa = 5.56QRR422 pKa = 11.84PHH424 pKa = 5.71PRR426 pKa = 11.84VTKK429 pKa = 9.66MM430 pKa = 3.37
MM1 pKa = 7.89DD2 pKa = 4.0KK3 pKa = 10.01WKK5 pKa = 9.21KK6 pKa = 9.18TIMHH10 pKa = 6.53MGAYY14 pKa = 8.22PAALGKK20 pKa = 9.24WVLLGGIIGVAGGVIGSLFHH40 pKa = 8.19IGVNYY45 pKa = 8.4ATAIRR50 pKa = 11.84GAHH53 pKa = 5.93PWILYY58 pKa = 7.62LLPVGGLAIVGLYY71 pKa = 9.65KK72 pKa = 10.54LCHH75 pKa = 6.72LEE77 pKa = 4.27GKK79 pKa = 8.16GTNAIIEE86 pKa = 4.44SVHH89 pKa = 6.84FGEE92 pKa = 5.05SVPILLVPVIFVSTVITHH110 pKa = 6.78LCGGSAGRR118 pKa = 11.84EE119 pKa = 3.93GAALQIGGGLGFQAGKK135 pKa = 10.35LLRR138 pKa = 11.84LGEE141 pKa = 4.34KK142 pKa = 10.13DD143 pKa = 4.13LPLATLCGMSGVFSALFGTPLTATVFALEE172 pKa = 4.38VISVGVLYY180 pKa = 10.84YY181 pKa = 10.89AGLVPCITAAMAAFGVSALMGVEE204 pKa = 3.98PTRR207 pKa = 11.84FTVSMPRR214 pKa = 11.84VTLEE218 pKa = 3.48LMLPVVVLSILCALVSILFCKK239 pKa = 9.44GLHH242 pKa = 4.59WTEE245 pKa = 4.12HH246 pKa = 5.85LLTRR250 pKa = 11.84SFRR253 pKa = 11.84NPWLRR258 pKa = 11.84VLAGAAILIVLSLLTNGDD276 pKa = 3.78YY277 pKa = 11.23NGAGMDD283 pKa = 3.86VIARR287 pKa = 11.84ALRR290 pKa = 11.84GNVSGWAWLGEE301 pKa = 4.21VLFFPLPPLLGAVHH315 pKa = 6.67RR316 pKa = 11.84RR317 pKa = 11.84HH318 pKa = 6.27HH319 pKa = 6.47RR320 pKa = 11.84LRR322 pKa = 11.84LQGRR326 pKa = 11.84RR327 pKa = 11.84GGAVVLRR334 pKa = 11.84GRR336 pKa = 11.84CLRR339 pKa = 11.84LLHH342 pKa = 6.62GGAPGPACGLCRR354 pKa = 11.84GHH356 pKa = 6.92RR357 pKa = 11.84PGGGVLRR364 pKa = 11.84GGEE367 pKa = 4.12LPRR370 pKa = 11.84GVGAAQRR377 pKa = 11.84GAVRR381 pKa = 11.84LCWGALLRR389 pKa = 11.84RR390 pKa = 11.84GLRR393 pKa = 11.84PEE395 pKa = 4.59LSPLRR400 pKa = 11.84LLRR403 pKa = 11.84PVQQPDD409 pKa = 3.25HH410 pKa = 7.12PLFQAPCGVHH420 pKa = 5.56QRR422 pKa = 11.84PHH424 pKa = 5.71PRR426 pKa = 11.84VTKK429 pKa = 9.66MM430 pKa = 3.37
Molecular weight: 45.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
571517 |
29 |
2348 |
313.5 |
34.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.834 ± 0.074 | 1.744 ± 0.031 |
5.832 ± 0.043 | 6.104 ± 0.062 |
3.469 ± 0.042 | 8.206 ± 0.105 |
1.993 ± 0.044 | 5.291 ± 0.048 |
4.265 ± 0.057 | 10.175 ± 0.076 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.837 ± 0.032 | 3.044 ± 0.04 |
4.233 ± 0.038 | 3.815 ± 0.04 |
6.394 ± 0.075 | 5.209 ± 0.05 |
5.531 ± 0.059 | 7.502 ± 0.062 |
1.042 ± 0.024 | 3.478 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
