
Merida virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Merhavirus; Merida merhavirus
Average proteome isoelectric point is 6.83
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
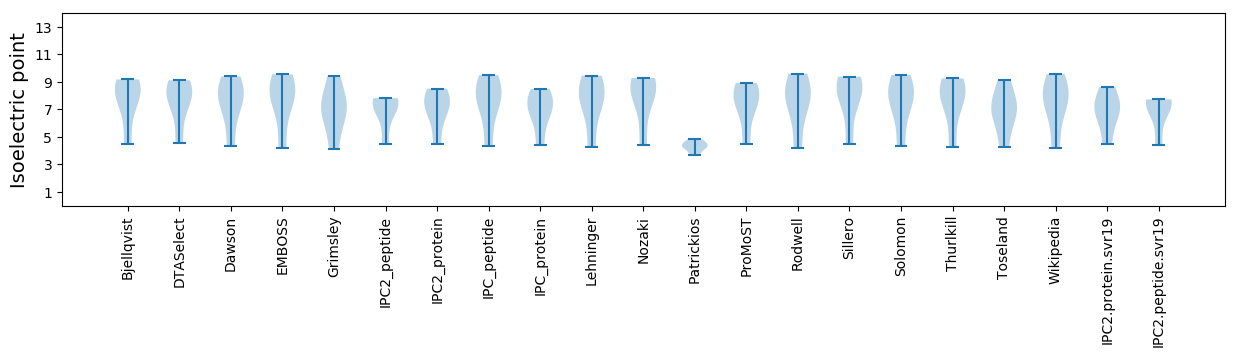
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140DDD9|A0A140DDD9_9RHAB Matrix protein OS=Merida virus OX=1803034 PE=4 SV=1
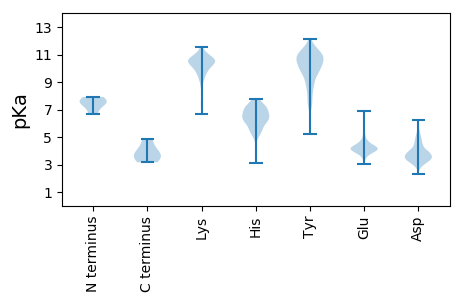
MM1 pKa = 7.45SNSGGEE7 pKa = 4.09SDD9 pKa = 4.32HH10 pKa = 5.53TTKK13 pKa = 10.65SQEE16 pKa = 3.85DD17 pKa = 3.53SRR19 pKa = 11.84IQGFYY24 pKa = 10.87SEE26 pKa = 5.07GVNLPQEE33 pKa = 4.5SVSCLGALLEE43 pKa = 4.08YY44 pKa = 10.6DD45 pKa = 3.71QEE47 pKa = 4.41GLEE50 pKa = 4.33KK51 pKa = 10.25TFSQMDD57 pKa = 3.88LEE59 pKa = 5.3DD60 pKa = 4.45PLTGEE65 pKa = 4.32RR66 pKa = 11.84EE67 pKa = 4.28GGSVQVSVPWGTVSEE82 pKa = 4.29EE83 pKa = 3.8QQAEE87 pKa = 4.3YY88 pKa = 10.98LRR90 pKa = 11.84LLRR93 pKa = 11.84LYY95 pKa = 9.87NIPHH99 pKa = 7.66PDD101 pKa = 3.44TPEE104 pKa = 3.75EE105 pKa = 4.02DD106 pKa = 3.26TGVDD110 pKa = 3.52EE111 pKa = 6.45DD112 pKa = 4.6EE113 pKa = 5.23LSSGTPSLPPTSPSLSSEE131 pKa = 3.93GGDD134 pKa = 3.15RR135 pKa = 11.84PLSPFLFSDD144 pKa = 3.94EE145 pKa = 4.85EE146 pKa = 4.07YY147 pKa = 10.74TEE149 pKa = 4.33YY150 pKa = 11.59VDD152 pKa = 3.11ITVPEE157 pKa = 4.12EE158 pKa = 4.04LQDD161 pKa = 3.61VALRR165 pKa = 11.84TPLISLLRR173 pKa = 11.84DD174 pKa = 3.4VVRR177 pKa = 11.84IMVNSKK183 pKa = 10.55NEE185 pKa = 3.98PFYY188 pKa = 11.33SLQKK192 pKa = 10.47YY193 pKa = 8.48DD194 pKa = 4.05LKK196 pKa = 11.12QGTVRR201 pKa = 11.84LARR204 pKa = 11.84VGNLYY209 pKa = 9.34TYY211 pKa = 10.09CPEE214 pKa = 4.07GTPIPVLEE222 pKa = 4.17QRR224 pKa = 11.84GSPKK228 pKa = 8.99TRR230 pKa = 11.84KK231 pKa = 9.27KK232 pKa = 10.55RR233 pKa = 11.84SLSTPQTNPTEE244 pKa = 4.69SSAQGPTSSGSRR256 pKa = 11.84ADD258 pKa = 3.71PTPSCSTTKK267 pKa = 10.45TDD269 pKa = 3.62IALGQTNQVSQTTEE283 pKa = 4.04EE284 pKa = 4.34SWPSPDD290 pKa = 4.37HH291 pKa = 6.84SGSPSNLEE299 pKa = 3.51GSVTIKK305 pKa = 10.46RR306 pKa = 11.84AASPSSSSHH315 pKa = 5.31YY316 pKa = 10.24SQTSSVEE323 pKa = 4.01SSSEE327 pKa = 3.81DD328 pKa = 3.37EE329 pKa = 4.46EE330 pKa = 4.61EE331 pKa = 4.31IEE333 pKa = 4.53VAQEE337 pKa = 4.52SYY339 pKa = 11.73SFMCPIYY346 pKa = 10.19GSSEE350 pKa = 3.75MSVKK354 pKa = 9.97VFPGHH359 pKa = 5.42EE360 pKa = 4.19VEE362 pKa = 5.12PLMKK366 pKa = 10.29LGMVPPEE373 pKa = 3.75IFRR376 pKa = 11.84AILRR380 pKa = 11.84DD381 pKa = 3.55RR382 pKa = 11.84GLLRR386 pKa = 11.84ALEE389 pKa = 4.2AVLDD393 pKa = 3.7IGQTHH398 pKa = 5.3MTT400 pKa = 3.99
MM1 pKa = 7.45SNSGGEE7 pKa = 4.09SDD9 pKa = 4.32HH10 pKa = 5.53TTKK13 pKa = 10.65SQEE16 pKa = 3.85DD17 pKa = 3.53SRR19 pKa = 11.84IQGFYY24 pKa = 10.87SEE26 pKa = 5.07GVNLPQEE33 pKa = 4.5SVSCLGALLEE43 pKa = 4.08YY44 pKa = 10.6DD45 pKa = 3.71QEE47 pKa = 4.41GLEE50 pKa = 4.33KK51 pKa = 10.25TFSQMDD57 pKa = 3.88LEE59 pKa = 5.3DD60 pKa = 4.45PLTGEE65 pKa = 4.32RR66 pKa = 11.84EE67 pKa = 4.28GGSVQVSVPWGTVSEE82 pKa = 4.29EE83 pKa = 3.8QQAEE87 pKa = 4.3YY88 pKa = 10.98LRR90 pKa = 11.84LLRR93 pKa = 11.84LYY95 pKa = 9.87NIPHH99 pKa = 7.66PDD101 pKa = 3.44TPEE104 pKa = 3.75EE105 pKa = 4.02DD106 pKa = 3.26TGVDD110 pKa = 3.52EE111 pKa = 6.45DD112 pKa = 4.6EE113 pKa = 5.23LSSGTPSLPPTSPSLSSEE131 pKa = 3.93GGDD134 pKa = 3.15RR135 pKa = 11.84PLSPFLFSDD144 pKa = 3.94EE145 pKa = 4.85EE146 pKa = 4.07YY147 pKa = 10.74TEE149 pKa = 4.33YY150 pKa = 11.59VDD152 pKa = 3.11ITVPEE157 pKa = 4.12EE158 pKa = 4.04LQDD161 pKa = 3.61VALRR165 pKa = 11.84TPLISLLRR173 pKa = 11.84DD174 pKa = 3.4VVRR177 pKa = 11.84IMVNSKK183 pKa = 10.55NEE185 pKa = 3.98PFYY188 pKa = 11.33SLQKK192 pKa = 10.47YY193 pKa = 8.48DD194 pKa = 4.05LKK196 pKa = 11.12QGTVRR201 pKa = 11.84LARR204 pKa = 11.84VGNLYY209 pKa = 9.34TYY211 pKa = 10.09CPEE214 pKa = 4.07GTPIPVLEE222 pKa = 4.17QRR224 pKa = 11.84GSPKK228 pKa = 8.99TRR230 pKa = 11.84KK231 pKa = 9.27KK232 pKa = 10.55RR233 pKa = 11.84SLSTPQTNPTEE244 pKa = 4.69SSAQGPTSSGSRR256 pKa = 11.84ADD258 pKa = 3.71PTPSCSTTKK267 pKa = 10.45TDD269 pKa = 3.62IALGQTNQVSQTTEE283 pKa = 4.04EE284 pKa = 4.34SWPSPDD290 pKa = 4.37HH291 pKa = 6.84SGSPSNLEE299 pKa = 3.51GSVTIKK305 pKa = 10.46RR306 pKa = 11.84AASPSSSSHH315 pKa = 5.31YY316 pKa = 10.24SQTSSVEE323 pKa = 4.01SSSEE327 pKa = 3.81DD328 pKa = 3.37EE329 pKa = 4.46EE330 pKa = 4.61EE331 pKa = 4.31IEE333 pKa = 4.53VAQEE337 pKa = 4.52SYY339 pKa = 11.73SFMCPIYY346 pKa = 10.19GSSEE350 pKa = 3.75MSVKK354 pKa = 9.97VFPGHH359 pKa = 5.42EE360 pKa = 4.19VEE362 pKa = 5.12PLMKK366 pKa = 10.29LGMVPPEE373 pKa = 3.75IFRR376 pKa = 11.84AILRR380 pKa = 11.84DD381 pKa = 3.55RR382 pKa = 11.84GLLRR386 pKa = 11.84ALEE389 pKa = 4.2AVLDD393 pKa = 3.7IGQTHH398 pKa = 5.3MTT400 pKa = 3.99
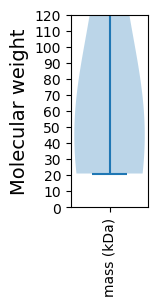
Molecular weight: 43.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140DDE0|A0A140DDE0_9RHAB Glycoprotein OS=Merida virus OX=1803034 PE=4 SV=1
MM1 pKa = 7.41SFVSMLKK8 pKa = 10.47GKK10 pKa = 9.26PGPSKK15 pKa = 10.43RR16 pKa = 11.84SPSTGHH22 pKa = 6.3VPFQGRR28 pKa = 11.84LDD30 pKa = 3.51MSLTIFTDD38 pKa = 3.33KK39 pKa = 10.27PYY41 pKa = 11.4SSMEE45 pKa = 3.8EE46 pKa = 3.96AFLHH50 pKa = 6.56LIEE53 pKa = 4.43IQDD56 pKa = 4.29YY57 pKa = 10.47ISEE60 pKa = 4.18TRR62 pKa = 11.84DD63 pKa = 3.34LKK65 pKa = 11.16GLYY68 pKa = 9.66LGLIAISILFSKK80 pKa = 10.67CDD82 pKa = 3.36ARR84 pKa = 11.84GNGVMYY90 pKa = 10.84TMDD93 pKa = 4.16LKK95 pKa = 11.33DD96 pKa = 4.11LVSLISTDD104 pKa = 3.13SALASGPLRR113 pKa = 11.84TFSYY117 pKa = 10.71EE118 pKa = 3.71RR119 pKa = 11.84AHH121 pKa = 6.97DD122 pKa = 4.12LRR124 pKa = 11.84GKK126 pKa = 7.5TALVRR131 pKa = 11.84FQLNFEE137 pKa = 4.16QSGLEE142 pKa = 4.18CEE144 pKa = 4.56SLLSVLLKK152 pKa = 10.02RR153 pKa = 11.84DD154 pKa = 3.23KK155 pKa = 11.07NKK157 pKa = 10.71EE158 pKa = 4.03VMSSSTSYY166 pKa = 11.12LKK168 pKa = 10.58LWRR171 pKa = 11.84LKK173 pKa = 10.39MIDD176 pKa = 3.84YY177 pKa = 10.71KK178 pKa = 10.92RR179 pKa = 11.84GTLFSLSS186 pKa = 3.5
MM1 pKa = 7.41SFVSMLKK8 pKa = 10.47GKK10 pKa = 9.26PGPSKK15 pKa = 10.43RR16 pKa = 11.84SPSTGHH22 pKa = 6.3VPFQGRR28 pKa = 11.84LDD30 pKa = 3.51MSLTIFTDD38 pKa = 3.33KK39 pKa = 10.27PYY41 pKa = 11.4SSMEE45 pKa = 3.8EE46 pKa = 3.96AFLHH50 pKa = 6.56LIEE53 pKa = 4.43IQDD56 pKa = 4.29YY57 pKa = 10.47ISEE60 pKa = 4.18TRR62 pKa = 11.84DD63 pKa = 3.34LKK65 pKa = 11.16GLYY68 pKa = 9.66LGLIAISILFSKK80 pKa = 10.67CDD82 pKa = 3.36ARR84 pKa = 11.84GNGVMYY90 pKa = 10.84TMDD93 pKa = 4.16LKK95 pKa = 11.33DD96 pKa = 4.11LVSLISTDD104 pKa = 3.13SALASGPLRR113 pKa = 11.84TFSYY117 pKa = 10.71EE118 pKa = 3.71RR119 pKa = 11.84AHH121 pKa = 6.97DD122 pKa = 4.12LRR124 pKa = 11.84GKK126 pKa = 7.5TALVRR131 pKa = 11.84FQLNFEE137 pKa = 4.16QSGLEE142 pKa = 4.18CEE144 pKa = 4.56SLLSVLLKK152 pKa = 10.02RR153 pKa = 11.84DD154 pKa = 3.23KK155 pKa = 11.07NKK157 pKa = 10.71EE158 pKa = 4.03VMSSSTSYY166 pKa = 11.12LKK168 pKa = 10.58LWRR171 pKa = 11.84LKK173 pKa = 10.39MIDD176 pKa = 3.84YY177 pKa = 10.71KK178 pKa = 10.92RR179 pKa = 11.84GTLFSLSS186 pKa = 3.5
Molecular weight: 20.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3708 |
186 |
2136 |
741.6 |
83.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.421 ± 0.487 | 1.591 ± 0.233 |
5.556 ± 0.372 | 6.311 ± 0.79 |
4.342 ± 0.707 | 7.012 ± 0.082 |
2.131 ± 0.145 | 5.771 ± 0.587 |
5.556 ± 0.642 | 10.086 ± 0.731 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.292 ± 0.287 | 3.209 ± 0.371 |
5.07 ± 0.713 | 3.344 ± 0.272 |
5.933 ± 0.458 | 9.736 ± 1.003 |
5.771 ± 0.56 | 5.69 ± 0.398 |
1.888 ± 0.387 | 3.29 ± 0.221 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
