
Batrachochytrium dendrobatidis (strain JEL423)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Chytridiomycota; Chytridiomycota incertae sedis; Chytridiomycetes; Rhizophydiales; Rhizophydiales incertae sedis; Batrachochytrium; Batrachochytrium dendrobatidis
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
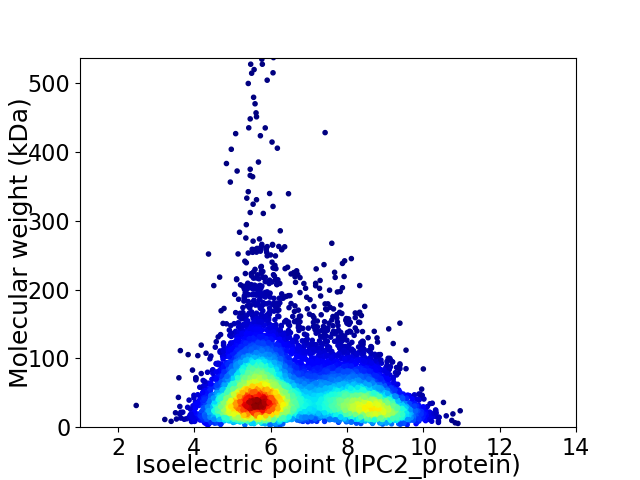
Virtual 2D-PAGE plot for 9531 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177X0J6|A0A177X0J6_BATDL BAG domain-containing protein OS=Batrachochytrium dendrobatidis (strain JEL423) OX=403673 GN=BDEG_28441 PE=4 SV=1
MM1 pKa = 7.42HH2 pKa = 7.85PSIQCQVDD10 pKa = 3.63TTSITKK16 pKa = 10.4NYY18 pKa = 9.28TSDD21 pKa = 3.64NIKK24 pKa = 10.85YY25 pKa = 9.71PDD27 pKa = 5.87DD28 pKa = 3.47IAQLDD33 pKa = 4.16DD34 pKa = 4.76EE35 pKa = 5.77LDD37 pKa = 3.82EE38 pKa = 4.47QDD40 pKa = 5.38SLFPDD45 pKa = 3.44ITILFNNTSYY55 pKa = 11.35PLFSTTAALVPDD67 pKa = 4.81DD68 pKa = 5.0YY69 pKa = 11.59IPFSFDD75 pKa = 3.86LDD77 pKa = 4.17EE78 pKa = 5.56EE79 pKa = 4.52DD80 pKa = 4.8LPSNVLLNSPLYY92 pKa = 10.17YY93 pKa = 10.47LVRR96 pKa = 11.84EE97 pKa = 4.46LKK99 pKa = 10.39HH100 pKa = 5.13VLNIKK105 pKa = 10.29SEE107 pKa = 4.17LYY109 pKa = 10.34DD110 pKa = 3.34VSIQIPQLDD119 pKa = 5.14LYY121 pKa = 11.52AMHH124 pKa = 7.06QDD126 pKa = 3.62LPILNRR132 pKa = 11.84LTLPTLIEE140 pKa = 3.89FHH142 pKa = 5.74QARR145 pKa = 11.84HH146 pKa = 4.7FQSVGSDD153 pKa = 3.18TASALPSLEE162 pKa = 4.82IILIQHH168 pKa = 5.77PTSLKK173 pKa = 9.78HH174 pKa = 6.0QLSLLEE180 pKa = 4.12QICTEE185 pKa = 4.47GGLDD189 pKa = 3.68SNPIVLDD196 pKa = 3.76SDD198 pKa = 4.03DD199 pKa = 5.66DD200 pKa = 4.21NLGDD204 pKa = 3.66EE205 pKa = 4.8YY206 pKa = 11.38EE207 pKa = 4.35NNTLALSTPIQPVSKK222 pKa = 10.41DD223 pKa = 3.09SHH225 pKa = 4.93FQKK228 pKa = 10.74NAAEE232 pKa = 4.36IDD234 pKa = 3.54IKK236 pKa = 11.32DD237 pKa = 3.97DD238 pKa = 3.99LLILGDD244 pKa = 4.35HH245 pKa = 6.45SLHH248 pKa = 6.97SDD250 pKa = 3.28GNLEE254 pKa = 4.04HH255 pKa = 6.53TASSVFTISTPGRR268 pKa = 11.84NHH270 pKa = 6.52TNTVVDD276 pKa = 3.57QTTEE280 pKa = 4.1DD281 pKa = 3.66EE282 pKa = 4.74NDD284 pKa = 3.36NNDD287 pKa = 5.36CIEE290 pKa = 3.82QDD292 pKa = 3.42NPKK295 pKa = 8.59TQIEE299 pKa = 4.36QTLSLNDD306 pKa = 3.35NHH308 pKa = 6.28SACLVTQISDD318 pKa = 3.58PTVEE322 pKa = 4.64AFISTPHH329 pKa = 5.94QPLSADD335 pKa = 3.08SSNPISTTEE344 pKa = 3.86CDD346 pKa = 3.39TTQVLPTDD354 pKa = 4.07SDD356 pKa = 4.55LVIQPVGIDD365 pKa = 3.32DD366 pKa = 4.22HH367 pKa = 8.79ASEE370 pKa = 5.54DD371 pKa = 3.87VLADD375 pKa = 3.33ASMIDD380 pKa = 4.47PEE382 pKa = 4.49ILDD385 pKa = 4.2AFDD388 pKa = 4.2SEE390 pKa = 4.89VVNDD394 pKa = 3.46EE395 pKa = 4.63MYY397 pKa = 11.23AEE399 pKa = 5.92DD400 pKa = 3.92IDD402 pKa = 4.46NDD404 pKa = 3.38EE405 pKa = 5.04TYY407 pKa = 10.66TEE409 pKa = 5.69DD410 pKa = 3.15IGNDD414 pKa = 2.86EE415 pKa = 4.82TYY417 pKa = 11.25AEE419 pKa = 5.29DD420 pKa = 3.26IGNDD424 pKa = 2.97EE425 pKa = 4.8TYY427 pKa = 11.25AEE429 pKa = 4.04DD430 pKa = 3.51TGNDD434 pKa = 3.17EE435 pKa = 6.03HH436 pKa = 8.66IDD438 pKa = 3.57ADD440 pKa = 3.83GSIAFTTFVSDD451 pKa = 3.53NTLQDD456 pKa = 2.74VDD458 pKa = 4.11STVLEE463 pKa = 3.95NTEE466 pKa = 3.76AHH468 pKa = 6.31FEE470 pKa = 4.42DD471 pKa = 5.05GFVAEE476 pKa = 4.25EE477 pKa = 4.17FEE479 pKa = 4.58YY480 pKa = 10.47TVQGDD485 pKa = 3.87EE486 pKa = 4.4STDD489 pKa = 3.47QYY491 pKa = 11.66SSNINDD497 pKa = 3.69DD498 pKa = 3.82TIDD501 pKa = 3.92AVDD504 pKa = 3.47STLIIDD510 pKa = 4.44LDD512 pKa = 3.77VAKK515 pKa = 10.87VVDD518 pKa = 3.81HH519 pKa = 6.65QGDD522 pKa = 3.48EE523 pKa = 4.4GTDD526 pKa = 3.39QVFDD530 pKa = 4.26DD531 pKa = 4.45PLVEE535 pKa = 4.25VEE537 pKa = 4.89LDD539 pKa = 3.62NMDD542 pKa = 4.88ALMDD546 pKa = 4.29DD547 pKa = 3.67MEE549 pKa = 4.5YY550 pKa = 10.41TEE552 pKa = 5.8EE553 pKa = 4.29IDD555 pKa = 3.58PHH557 pKa = 7.68VDD559 pKa = 3.39EE560 pKa = 5.72NDD562 pKa = 3.09VALDD566 pKa = 3.35VDD568 pKa = 4.15TDD570 pKa = 3.86HH571 pKa = 7.66QSLQNTDD578 pKa = 3.28QAKK581 pKa = 10.57DD582 pKa = 3.33SADD585 pKa = 3.12AHH587 pKa = 6.37ILLEE591 pKa = 4.48KK592 pKa = 9.87ATADD596 pKa = 4.43AIHH599 pKa = 6.92DD600 pKa = 3.83TALDD604 pKa = 3.89AVCIDD609 pKa = 3.89PTNSNLDD616 pKa = 3.71SEE618 pKa = 5.11VDD620 pKa = 3.52DD621 pKa = 4.31LNAYY625 pKa = 9.31KK626 pKa = 10.54FSLGSKK632 pKa = 9.91RR633 pKa = 11.84EE634 pKa = 4.07LEE636 pKa = 4.19QEE638 pKa = 4.16HH639 pKa = 7.52DD640 pKa = 3.49DD641 pKa = 4.02AAAKK645 pKa = 9.81RR646 pKa = 11.84ARR648 pKa = 11.84VV649 pKa = 3.34
MM1 pKa = 7.42HH2 pKa = 7.85PSIQCQVDD10 pKa = 3.63TTSITKK16 pKa = 10.4NYY18 pKa = 9.28TSDD21 pKa = 3.64NIKK24 pKa = 10.85YY25 pKa = 9.71PDD27 pKa = 5.87DD28 pKa = 3.47IAQLDD33 pKa = 4.16DD34 pKa = 4.76EE35 pKa = 5.77LDD37 pKa = 3.82EE38 pKa = 4.47QDD40 pKa = 5.38SLFPDD45 pKa = 3.44ITILFNNTSYY55 pKa = 11.35PLFSTTAALVPDD67 pKa = 4.81DD68 pKa = 5.0YY69 pKa = 11.59IPFSFDD75 pKa = 3.86LDD77 pKa = 4.17EE78 pKa = 5.56EE79 pKa = 4.52DD80 pKa = 4.8LPSNVLLNSPLYY92 pKa = 10.17YY93 pKa = 10.47LVRR96 pKa = 11.84EE97 pKa = 4.46LKK99 pKa = 10.39HH100 pKa = 5.13VLNIKK105 pKa = 10.29SEE107 pKa = 4.17LYY109 pKa = 10.34DD110 pKa = 3.34VSIQIPQLDD119 pKa = 5.14LYY121 pKa = 11.52AMHH124 pKa = 7.06QDD126 pKa = 3.62LPILNRR132 pKa = 11.84LTLPTLIEE140 pKa = 3.89FHH142 pKa = 5.74QARR145 pKa = 11.84HH146 pKa = 4.7FQSVGSDD153 pKa = 3.18TASALPSLEE162 pKa = 4.82IILIQHH168 pKa = 5.77PTSLKK173 pKa = 9.78HH174 pKa = 6.0QLSLLEE180 pKa = 4.12QICTEE185 pKa = 4.47GGLDD189 pKa = 3.68SNPIVLDD196 pKa = 3.76SDD198 pKa = 4.03DD199 pKa = 5.66DD200 pKa = 4.21NLGDD204 pKa = 3.66EE205 pKa = 4.8YY206 pKa = 11.38EE207 pKa = 4.35NNTLALSTPIQPVSKK222 pKa = 10.41DD223 pKa = 3.09SHH225 pKa = 4.93FQKK228 pKa = 10.74NAAEE232 pKa = 4.36IDD234 pKa = 3.54IKK236 pKa = 11.32DD237 pKa = 3.97DD238 pKa = 3.99LLILGDD244 pKa = 4.35HH245 pKa = 6.45SLHH248 pKa = 6.97SDD250 pKa = 3.28GNLEE254 pKa = 4.04HH255 pKa = 6.53TASSVFTISTPGRR268 pKa = 11.84NHH270 pKa = 6.52TNTVVDD276 pKa = 3.57QTTEE280 pKa = 4.1DD281 pKa = 3.66EE282 pKa = 4.74NDD284 pKa = 3.36NNDD287 pKa = 5.36CIEE290 pKa = 3.82QDD292 pKa = 3.42NPKK295 pKa = 8.59TQIEE299 pKa = 4.36QTLSLNDD306 pKa = 3.35NHH308 pKa = 6.28SACLVTQISDD318 pKa = 3.58PTVEE322 pKa = 4.64AFISTPHH329 pKa = 5.94QPLSADD335 pKa = 3.08SSNPISTTEE344 pKa = 3.86CDD346 pKa = 3.39TTQVLPTDD354 pKa = 4.07SDD356 pKa = 4.55LVIQPVGIDD365 pKa = 3.32DD366 pKa = 4.22HH367 pKa = 8.79ASEE370 pKa = 5.54DD371 pKa = 3.87VLADD375 pKa = 3.33ASMIDD380 pKa = 4.47PEE382 pKa = 4.49ILDD385 pKa = 4.2AFDD388 pKa = 4.2SEE390 pKa = 4.89VVNDD394 pKa = 3.46EE395 pKa = 4.63MYY397 pKa = 11.23AEE399 pKa = 5.92DD400 pKa = 3.92IDD402 pKa = 4.46NDD404 pKa = 3.38EE405 pKa = 5.04TYY407 pKa = 10.66TEE409 pKa = 5.69DD410 pKa = 3.15IGNDD414 pKa = 2.86EE415 pKa = 4.82TYY417 pKa = 11.25AEE419 pKa = 5.29DD420 pKa = 3.26IGNDD424 pKa = 2.97EE425 pKa = 4.8TYY427 pKa = 11.25AEE429 pKa = 4.04DD430 pKa = 3.51TGNDD434 pKa = 3.17EE435 pKa = 6.03HH436 pKa = 8.66IDD438 pKa = 3.57ADD440 pKa = 3.83GSIAFTTFVSDD451 pKa = 3.53NTLQDD456 pKa = 2.74VDD458 pKa = 4.11STVLEE463 pKa = 3.95NTEE466 pKa = 3.76AHH468 pKa = 6.31FEE470 pKa = 4.42DD471 pKa = 5.05GFVAEE476 pKa = 4.25EE477 pKa = 4.17FEE479 pKa = 4.58YY480 pKa = 10.47TVQGDD485 pKa = 3.87EE486 pKa = 4.4STDD489 pKa = 3.47QYY491 pKa = 11.66SSNINDD497 pKa = 3.69DD498 pKa = 3.82TIDD501 pKa = 3.92AVDD504 pKa = 3.47STLIIDD510 pKa = 4.44LDD512 pKa = 3.77VAKK515 pKa = 10.87VVDD518 pKa = 3.81HH519 pKa = 6.65QGDD522 pKa = 3.48EE523 pKa = 4.4GTDD526 pKa = 3.39QVFDD530 pKa = 4.26DD531 pKa = 4.45PLVEE535 pKa = 4.25VEE537 pKa = 4.89LDD539 pKa = 3.62NMDD542 pKa = 4.88ALMDD546 pKa = 4.29DD547 pKa = 3.67MEE549 pKa = 4.5YY550 pKa = 10.41TEE552 pKa = 5.8EE553 pKa = 4.29IDD555 pKa = 3.58PHH557 pKa = 7.68VDD559 pKa = 3.39EE560 pKa = 5.72NDD562 pKa = 3.09VALDD566 pKa = 3.35VDD568 pKa = 4.15TDD570 pKa = 3.86HH571 pKa = 7.66QSLQNTDD578 pKa = 3.28QAKK581 pKa = 10.57DD582 pKa = 3.33SADD585 pKa = 3.12AHH587 pKa = 6.37ILLEE591 pKa = 4.48KK592 pKa = 9.87ATADD596 pKa = 4.43AIHH599 pKa = 6.92DD600 pKa = 3.83TALDD604 pKa = 3.89AVCIDD609 pKa = 3.89PTNSNLDD616 pKa = 3.71SEE618 pKa = 5.11VDD620 pKa = 3.52DD621 pKa = 4.31LNAYY625 pKa = 9.31KK626 pKa = 10.54FSLGSKK632 pKa = 9.91RR633 pKa = 11.84EE634 pKa = 4.07LEE636 pKa = 4.19QEE638 pKa = 4.16HH639 pKa = 7.52DD640 pKa = 3.49DD641 pKa = 4.02AAAKK645 pKa = 9.81RR646 pKa = 11.84ARR648 pKa = 11.84VV649 pKa = 3.34
Molecular weight: 71.87 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177WM73|A0A177WM73_BATDL Uncharacterized protein OS=Batrachochytrium dendrobatidis (strain JEL423) OX=403673 GN=BDEG_24852 PE=3 SV=1
MM1 pKa = 7.07PHH3 pKa = 6.59SRR5 pKa = 11.84LLCVFSSNAFKK16 pKa = 11.05LPLPNVRR23 pKa = 11.84EE24 pKa = 4.27KK25 pKa = 10.69IVLPLIRR32 pKa = 11.84LQHH35 pKa = 6.59PPLQPALDD43 pKa = 3.77LTALQRR49 pKa = 11.84KK50 pKa = 7.82LPWMLTMRR58 pKa = 11.84RR59 pKa = 11.84IQMPMVKK66 pKa = 9.23ISKK69 pKa = 8.34MNSMNLMTKK78 pKa = 9.9ISRR81 pKa = 11.84KK82 pKa = 8.18GTSLPKK88 pKa = 9.82PLPPRR93 pKa = 11.84MTIKK97 pKa = 9.52STRR100 pKa = 11.84IPSLLPRR107 pKa = 11.84VKK109 pKa = 9.52STRR112 pKa = 11.84IPNPLPRR119 pKa = 11.84LEE121 pKa = 4.16VKK123 pKa = 9.64RR124 pKa = 11.84RR125 pKa = 11.84KK126 pKa = 9.69RR127 pKa = 11.84MRR129 pKa = 11.84RR130 pKa = 11.84KK131 pKa = 7.03MRR133 pKa = 11.84KK134 pKa = 8.06RR135 pKa = 11.84IRR137 pKa = 11.84SIRR140 pKa = 11.84IPSLLPSSPRR150 pKa = 11.84LQLLPRR156 pKa = 11.84PLPRR160 pKa = 11.84PQLLPRR166 pKa = 11.84LLLLPRR172 pKa = 11.84PQLPPRR178 pKa = 11.84PLQRR182 pKa = 11.84PQPRR186 pKa = 11.84PLLQPLPLQVKK197 pKa = 9.89ASTLRR202 pKa = 11.84RR203 pKa = 11.84MM204 pKa = 3.95
MM1 pKa = 7.07PHH3 pKa = 6.59SRR5 pKa = 11.84LLCVFSSNAFKK16 pKa = 11.05LPLPNVRR23 pKa = 11.84EE24 pKa = 4.27KK25 pKa = 10.69IVLPLIRR32 pKa = 11.84LQHH35 pKa = 6.59PPLQPALDD43 pKa = 3.77LTALQRR49 pKa = 11.84KK50 pKa = 7.82LPWMLTMRR58 pKa = 11.84RR59 pKa = 11.84IQMPMVKK66 pKa = 9.23ISKK69 pKa = 8.34MNSMNLMTKK78 pKa = 9.9ISRR81 pKa = 11.84KK82 pKa = 8.18GTSLPKK88 pKa = 9.82PLPPRR93 pKa = 11.84MTIKK97 pKa = 9.52STRR100 pKa = 11.84IPSLLPRR107 pKa = 11.84VKK109 pKa = 9.52STRR112 pKa = 11.84IPNPLPRR119 pKa = 11.84LEE121 pKa = 4.16VKK123 pKa = 9.64RR124 pKa = 11.84RR125 pKa = 11.84KK126 pKa = 9.69RR127 pKa = 11.84MRR129 pKa = 11.84RR130 pKa = 11.84KK131 pKa = 7.03MRR133 pKa = 11.84KK134 pKa = 8.06RR135 pKa = 11.84IRR137 pKa = 11.84SIRR140 pKa = 11.84IPSLLPSSPRR150 pKa = 11.84LQLLPRR156 pKa = 11.84PLPRR160 pKa = 11.84PQLLPRR166 pKa = 11.84LLLLPRR172 pKa = 11.84PQLPPRR178 pKa = 11.84PLQRR182 pKa = 11.84PQPRR186 pKa = 11.84PLLQPLPLQVKK197 pKa = 9.89ASTLRR202 pKa = 11.84RR203 pKa = 11.84MM204 pKa = 3.95
Molecular weight: 23.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4695730 |
29 |
4754 |
492.7 |
54.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.013 ± 0.022 | 1.47 ± 0.01 |
5.698 ± 0.017 | 5.392 ± 0.024 |
3.818 ± 0.014 | 5.086 ± 0.022 |
2.748 ± 0.013 | 6.163 ± 0.016 |
5.701 ± 0.026 | 9.258 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.437 ± 0.009 | 4.609 ± 0.014 |
4.988 ± 0.026 | 4.742 ± 0.02 |
4.759 ± 0.017 | 9.765 ± 0.031 |
6.558 ± 0.02 | 5.98 ± 0.018 |
1.005 ± 0.007 | 2.813 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
