
Myocastor coypus polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
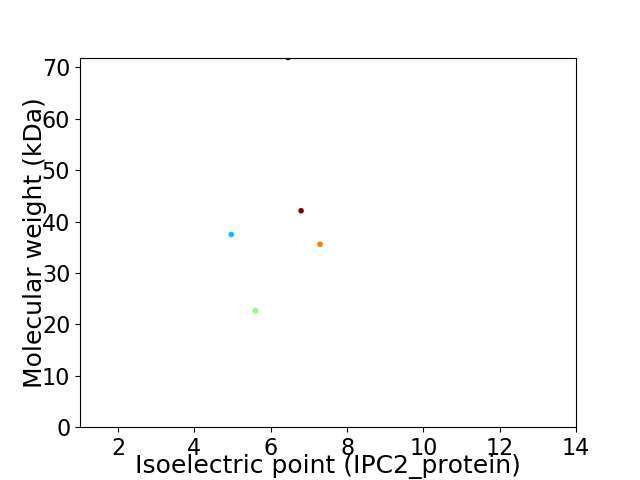
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346RRK3|A0A346RRK3_9POLY Minor capsid protein VP2 OS=Myocastor coypus polyomavirus 1 OX=2304056 PE=3 SV=1
MM1 pKa = 7.19GAVLAVIAEE10 pKa = 4.2VFGFEE15 pKa = 4.07LAAAGTAGATATALAEE31 pKa = 4.53LGTEE35 pKa = 4.17TVLTGEE41 pKa = 4.84LVTSLEE47 pKa = 4.13SQIASIMAIEE57 pKa = 4.98GVTQTEE63 pKa = 3.87ALAALGISAEE73 pKa = 4.35SYY75 pKa = 10.29GALTEE80 pKa = 4.72AYY82 pKa = 10.01LAGVDD87 pKa = 4.6ALGTQIGSGVSTGLGQTLIQGLSNINVPIGPFFQTITGTSYY128 pKa = 11.14LVAAGIQFHH137 pKa = 6.06KK138 pKa = 10.31HH139 pKa = 3.73QVSIVNRR146 pKa = 11.84NNMALVPYY154 pKa = 10.05IPEE157 pKa = 4.57DD158 pKa = 3.77YY159 pKa = 11.11YY160 pKa = 11.44DD161 pKa = 3.39ILFPGVQSFTRR172 pKa = 11.84GLNVVEE178 pKa = 4.75NWATSLFRR186 pKa = 11.84SLGHH190 pKa = 6.31YY191 pKa = 9.68LFQQLVRR198 pKa = 11.84EE199 pKa = 4.29AHH201 pKa = 5.48AQIGQAGRR209 pKa = 11.84SAGNALISYY218 pKa = 7.01GTRR221 pKa = 11.84GLEE224 pKa = 3.62EE225 pKa = 3.88SVARR229 pKa = 11.84FLEE232 pKa = 4.2SARR235 pKa = 11.84WVIQGPTNTYY245 pKa = 10.13NFLRR249 pKa = 11.84QYY251 pKa = 10.97YY252 pKa = 10.37EE253 pKa = 3.77NLTPLNPPRR262 pKa = 11.84FVQLEE267 pKa = 3.78KK268 pKa = 10.69RR269 pKa = 11.84LRR271 pKa = 11.84YY272 pKa = 9.52LDD274 pKa = 3.79QVPKK278 pKa = 10.15RR279 pKa = 11.84QAPGEE284 pKa = 4.13SGEE287 pKa = 4.26IVDD290 pKa = 4.66TVSSPGGALQRR301 pKa = 11.84HH302 pKa = 5.33TPDD305 pKa = 2.44WMLPLILGLYY315 pKa = 10.31GDD317 pKa = 5.09ISPTWRR323 pKa = 11.84TYY325 pKa = 10.35IEE327 pKa = 4.19EE328 pKa = 4.18EE329 pKa = 4.15EE330 pKa = 4.21QEE332 pKa = 4.15EE333 pKa = 4.95EE334 pKa = 4.13YY335 pKa = 11.17GPKK338 pKa = 9.86KK339 pKa = 10.41KK340 pKa = 9.95RR341 pKa = 11.84VRR343 pKa = 11.84YY344 pKa = 9.27DD345 pKa = 2.82
MM1 pKa = 7.19GAVLAVIAEE10 pKa = 4.2VFGFEE15 pKa = 4.07LAAAGTAGATATALAEE31 pKa = 4.53LGTEE35 pKa = 4.17TVLTGEE41 pKa = 4.84LVTSLEE47 pKa = 4.13SQIASIMAIEE57 pKa = 4.98GVTQTEE63 pKa = 3.87ALAALGISAEE73 pKa = 4.35SYY75 pKa = 10.29GALTEE80 pKa = 4.72AYY82 pKa = 10.01LAGVDD87 pKa = 4.6ALGTQIGSGVSTGLGQTLIQGLSNINVPIGPFFQTITGTSYY128 pKa = 11.14LVAAGIQFHH137 pKa = 6.06KK138 pKa = 10.31HH139 pKa = 3.73QVSIVNRR146 pKa = 11.84NNMALVPYY154 pKa = 10.05IPEE157 pKa = 4.57DD158 pKa = 3.77YY159 pKa = 11.11YY160 pKa = 11.44DD161 pKa = 3.39ILFPGVQSFTRR172 pKa = 11.84GLNVVEE178 pKa = 4.75NWATSLFRR186 pKa = 11.84SLGHH190 pKa = 6.31YY191 pKa = 9.68LFQQLVRR198 pKa = 11.84EE199 pKa = 4.29AHH201 pKa = 5.48AQIGQAGRR209 pKa = 11.84SAGNALISYY218 pKa = 7.01GTRR221 pKa = 11.84GLEE224 pKa = 3.62EE225 pKa = 3.88SVARR229 pKa = 11.84FLEE232 pKa = 4.2SARR235 pKa = 11.84WVIQGPTNTYY245 pKa = 10.13NFLRR249 pKa = 11.84QYY251 pKa = 10.97YY252 pKa = 10.37EE253 pKa = 3.77NLTPLNPPRR262 pKa = 11.84FVQLEE267 pKa = 3.78KK268 pKa = 10.69RR269 pKa = 11.84LRR271 pKa = 11.84YY272 pKa = 9.52LDD274 pKa = 3.79QVPKK278 pKa = 10.15RR279 pKa = 11.84QAPGEE284 pKa = 4.13SGEE287 pKa = 4.26IVDD290 pKa = 4.66TVSSPGGALQRR301 pKa = 11.84HH302 pKa = 5.33TPDD305 pKa = 2.44WMLPLILGLYY315 pKa = 10.31GDD317 pKa = 5.09ISPTWRR323 pKa = 11.84TYY325 pKa = 10.35IEE327 pKa = 4.19EE328 pKa = 4.18EE329 pKa = 4.15EE330 pKa = 4.21QEE332 pKa = 4.15EE333 pKa = 4.95EE334 pKa = 4.13YY335 pKa = 11.17GPKK338 pKa = 9.86KK339 pKa = 10.41KK340 pKa = 9.95RR341 pKa = 11.84VRR343 pKa = 11.84YY344 pKa = 9.27DD345 pKa = 2.82
Molecular weight: 37.48 kDa
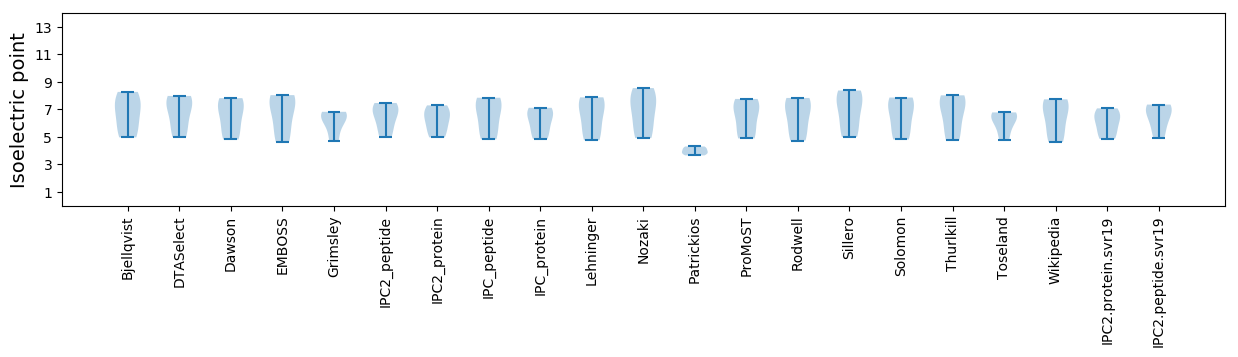
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346RRK5|A0A346RRK5_9POLY Large T antigen OS=Myocastor coypus polyomavirus 1 OX=2304056 PE=4 SV=1
MM1 pKa = 7.84DD2 pKa = 5.23RR3 pKa = 11.84VLGWGEE9 pKa = 4.25RR10 pKa = 11.84KK11 pKa = 9.78RR12 pKa = 11.84LMHH15 pKa = 6.82LLGVPLEE22 pKa = 4.47KK23 pKa = 10.81YY24 pKa = 10.51GNYY27 pKa = 9.94PVMRR31 pKa = 11.84DD32 pKa = 3.31AYY34 pKa = 9.01KK35 pKa = 10.81SKK37 pKa = 10.25VLEE40 pKa = 3.9YY41 pKa = 10.65HH42 pKa = 6.81PDD44 pKa = 3.22KK45 pKa = 11.63GGDD48 pKa = 3.39GKK50 pKa = 11.49LMSEE54 pKa = 5.6LNNLWKK60 pKa = 10.8AFTDD64 pKa = 4.07GLHH67 pKa = 7.09LLRR70 pKa = 11.84EE71 pKa = 4.36CEE73 pKa = 4.12EE74 pKa = 4.19VQKK77 pKa = 10.85QGYY80 pKa = 7.59TDD82 pKa = 4.71DD83 pKa = 4.15DD84 pKa = 4.18TVKK87 pKa = 10.73LFLGTPIKK95 pKa = 10.58GVYY98 pKa = 9.43CKK100 pKa = 10.59VYY102 pKa = 9.72PLCKK106 pKa = 10.31DD107 pKa = 3.83FLHH110 pKa = 6.69KK111 pKa = 10.21KK112 pKa = 8.7CKK114 pKa = 10.4CIICVLKK121 pKa = 10.14RR122 pKa = 11.84QHH124 pKa = 6.13YY125 pKa = 9.05RR126 pKa = 11.84MKK128 pKa = 10.93ARR130 pKa = 11.84TLKK133 pKa = 10.63LCLMWGQCYY142 pKa = 10.03CYY144 pKa = 9.99HH145 pKa = 7.74CYY147 pKa = 10.28LQWFGEE153 pKa = 4.0DD154 pKa = 3.97DD155 pKa = 3.9SYY157 pKa = 11.78DD158 pKa = 2.96ICKK161 pKa = 10.23RR162 pKa = 11.84YY163 pKa = 10.52LEE165 pKa = 4.79TIILMPINLIIYY177 pKa = 9.73DD178 pKa = 4.21SIFEE182 pKa = 4.09CEE184 pKa = 3.94EE185 pKa = 3.54WSKK188 pKa = 11.44YY189 pKa = 10.48LISLRR194 pKa = 11.84SIDD197 pKa = 4.66LYY199 pKa = 10.82ISLKK203 pKa = 9.8VLCDD207 pKa = 3.41CLDD210 pKa = 3.71RR211 pKa = 11.84KK212 pKa = 8.83QTVAEE217 pKa = 4.35APHH220 pKa = 6.23MEE222 pKa = 4.54LKK224 pKa = 10.54PSEE227 pKa = 4.17EE228 pKa = 4.39HH229 pKa = 6.8GMQLGEE235 pKa = 4.77LEE237 pKa = 4.3ALQARR242 pKa = 11.84LNPTPTQTSFVMNPWVSPPLMTSTPRR268 pKa = 11.84LNRR271 pKa = 11.84HH272 pKa = 5.58LHH274 pKa = 5.71LAPPRR279 pKa = 11.84GEE281 pKa = 4.37EE282 pKa = 4.03GDD284 pKa = 3.65KK285 pKa = 11.11ARR287 pKa = 11.84SNPTPTKK294 pKa = 9.88TFFGLTPGGQPPCC307 pKa = 4.38
MM1 pKa = 7.84DD2 pKa = 5.23RR3 pKa = 11.84VLGWGEE9 pKa = 4.25RR10 pKa = 11.84KK11 pKa = 9.78RR12 pKa = 11.84LMHH15 pKa = 6.82LLGVPLEE22 pKa = 4.47KK23 pKa = 10.81YY24 pKa = 10.51GNYY27 pKa = 9.94PVMRR31 pKa = 11.84DD32 pKa = 3.31AYY34 pKa = 9.01KK35 pKa = 10.81SKK37 pKa = 10.25VLEE40 pKa = 3.9YY41 pKa = 10.65HH42 pKa = 6.81PDD44 pKa = 3.22KK45 pKa = 11.63GGDD48 pKa = 3.39GKK50 pKa = 11.49LMSEE54 pKa = 5.6LNNLWKK60 pKa = 10.8AFTDD64 pKa = 4.07GLHH67 pKa = 7.09LLRR70 pKa = 11.84EE71 pKa = 4.36CEE73 pKa = 4.12EE74 pKa = 4.19VQKK77 pKa = 10.85QGYY80 pKa = 7.59TDD82 pKa = 4.71DD83 pKa = 4.15DD84 pKa = 4.18TVKK87 pKa = 10.73LFLGTPIKK95 pKa = 10.58GVYY98 pKa = 9.43CKK100 pKa = 10.59VYY102 pKa = 9.72PLCKK106 pKa = 10.31DD107 pKa = 3.83FLHH110 pKa = 6.69KK111 pKa = 10.21KK112 pKa = 8.7CKK114 pKa = 10.4CIICVLKK121 pKa = 10.14RR122 pKa = 11.84QHH124 pKa = 6.13YY125 pKa = 9.05RR126 pKa = 11.84MKK128 pKa = 10.93ARR130 pKa = 11.84TLKK133 pKa = 10.63LCLMWGQCYY142 pKa = 10.03CYY144 pKa = 9.99HH145 pKa = 7.74CYY147 pKa = 10.28LQWFGEE153 pKa = 4.0DD154 pKa = 3.97DD155 pKa = 3.9SYY157 pKa = 11.78DD158 pKa = 2.96ICKK161 pKa = 10.23RR162 pKa = 11.84YY163 pKa = 10.52LEE165 pKa = 4.79TIILMPINLIIYY177 pKa = 9.73DD178 pKa = 4.21SIFEE182 pKa = 4.09CEE184 pKa = 3.94EE185 pKa = 3.54WSKK188 pKa = 11.44YY189 pKa = 10.48LISLRR194 pKa = 11.84SIDD197 pKa = 4.66LYY199 pKa = 10.82ISLKK203 pKa = 9.8VLCDD207 pKa = 3.41CLDD210 pKa = 3.71RR211 pKa = 11.84KK212 pKa = 8.83QTVAEE217 pKa = 4.35APHH220 pKa = 6.23MEE222 pKa = 4.54LKK224 pKa = 10.54PSEE227 pKa = 4.17EE228 pKa = 4.39HH229 pKa = 6.8GMQLGEE235 pKa = 4.77LEE237 pKa = 4.3ALQARR242 pKa = 11.84LNPTPTQTSFVMNPWVSPPLMTSTPRR268 pKa = 11.84LNRR271 pKa = 11.84HH272 pKa = 5.58LHH274 pKa = 5.71LAPPRR279 pKa = 11.84GEE281 pKa = 4.37EE282 pKa = 4.03GDD284 pKa = 3.65KK285 pKa = 11.11ARR287 pKa = 11.84SNPTPTKK294 pKa = 9.88TFFGLTPGGQPPCC307 pKa = 4.38
Molecular weight: 35.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1857 |
197 |
625 |
371.4 |
41.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.331 ± 1.143 | 2.423 ± 0.889 |
4.362 ± 0.604 | 7.701 ± 0.342 |
3.446 ± 0.365 | 7.485 ± 1.026 |
2.154 ± 0.465 | 5.17 ± 0.29 |
6.354 ± 1.488 | 10.232 ± 1.013 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.477 ± 0.4 | 3.823 ± 0.415 |
5.924 ± 0.717 | 4.47 ± 0.432 |
5.116 ± 0.507 | 5.6 ± 0.373 |
5.87 ± 0.771 | 6.462 ± 0.601 |
1.4 ± 0.192 | 4.2 ± 0.608 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
