
SAR86 cluster bacterium SAR86B
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; SAR86 cluster
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
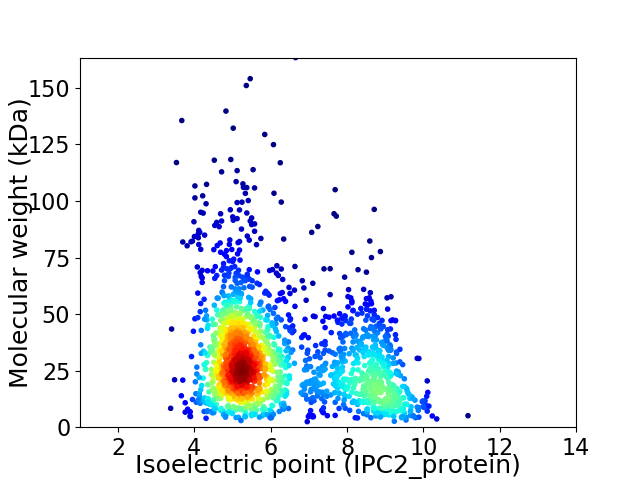
Virtual 2D-PAGE plot for 1807 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J4WX98|J4WX98_9GAMM Uncharacterized protein OS=SAR86 cluster bacterium SAR86B OX=1123867 GN=NT02SARS_1192 PE=4 SV=1
MM1 pKa = 7.51KK2 pKa = 10.29SKK4 pKa = 10.89SSLFIAFLFLFSCGGGGGGGSAALPLPIITIAASSLDD41 pKa = 3.82VNIGDD46 pKa = 4.94DD47 pKa = 4.75VDD49 pKa = 4.06ITWSVSNSNSCNATGAWSGIKK70 pKa = 10.19SNSGNEE76 pKa = 4.05TVTVSGAGNNNFGLSCSGEE95 pKa = 4.04GGQASEE101 pKa = 4.46TVTVFAFNLAANNTTLAVDD120 pKa = 3.82EE121 pKa = 5.47DD122 pKa = 4.29GSLINSSVAVQPNTDD137 pKa = 2.99ILVSYY142 pKa = 10.36SLLSSTSNGNLTFNEE157 pKa = 4.39DD158 pKa = 2.91ATINYY163 pKa = 9.29YY164 pKa = 10.0PDD166 pKa = 3.85PNFNGVDD173 pKa = 3.25EE174 pKa = 4.84FTFKK178 pKa = 10.96AIAATRR184 pKa = 11.84SVEE187 pKa = 3.83KK188 pKa = 10.8DD189 pKa = 3.05VTVTINIASINDD201 pKa = 3.57APTIALDD208 pKa = 4.02SEE210 pKa = 5.11SNLSKK215 pKa = 11.0LDD217 pKa = 3.43MVYY220 pKa = 10.72EE221 pKa = 4.18SDD223 pKa = 4.94PIFSFNFADD232 pKa = 3.72VDD234 pKa = 3.88HH235 pKa = 6.95LLEE238 pKa = 5.19DD239 pKa = 3.75LTFTASVDD247 pKa = 3.54GNAVPTTFNQISEE260 pKa = 5.0GYY262 pKa = 10.65GSLQLDD268 pKa = 4.5LSSLGSGGLFDD279 pKa = 4.26LTLSVFDD286 pKa = 4.76GFDD289 pKa = 3.25TATEE293 pKa = 4.12TLNTWFVADD302 pKa = 4.0KK303 pKa = 10.47STVTFDD309 pKa = 4.43QDD311 pKa = 3.93DD312 pKa = 4.09DD313 pKa = 4.88PEE315 pKa = 6.54DD316 pKa = 4.0GVTEE320 pKa = 4.55GSSTSVDD327 pKa = 3.35YY328 pKa = 11.07NVYY331 pKa = 10.24YY332 pKa = 11.02LDD334 pKa = 4.42GNSEE338 pKa = 4.08SRR340 pKa = 11.84GRR342 pKa = 11.84TTYY345 pKa = 11.25LFVADD350 pKa = 4.23SLASEE355 pKa = 4.47EE356 pKa = 4.34DD357 pKa = 3.33RR358 pKa = 11.84ASFRR362 pKa = 11.84SALVRR367 pKa = 11.84SINKK371 pKa = 9.25VKK373 pKa = 10.63EE374 pKa = 3.78SDD376 pKa = 3.1AGEE379 pKa = 4.58FITGFFTIKK388 pKa = 10.16AAEE391 pKa = 3.92PVIPDD396 pKa = 3.91GKK398 pKa = 10.33SPSAIRR404 pKa = 11.84TGCYY408 pKa = 10.39DD409 pKa = 4.32FDD411 pKa = 4.97PNIYY415 pKa = 10.28CIGDD419 pKa = 3.38MDD421 pKa = 4.12TSVFDD426 pKa = 3.46VMYY429 pKa = 9.65PGHH432 pKa = 7.12LLVSTLTMQSGRR444 pKa = 11.84GVNLGNRR451 pKa = 11.84NIQPISSRR459 pKa = 11.84TQNVLMHH466 pKa = 6.52EE467 pKa = 4.86LGHH470 pKa = 6.39AHH472 pKa = 6.91GFMGDD477 pKa = 3.69EE478 pKa = 4.24YY479 pKa = 11.11RR480 pKa = 11.84SDD482 pKa = 3.9DD483 pKa = 4.79DD484 pKa = 5.68RR485 pKa = 11.84DD486 pKa = 3.32VSYY489 pKa = 9.81WADD492 pKa = 3.47LNINTTTQSDD502 pKa = 3.81PALVKK507 pKa = 9.65WKK509 pKa = 10.26HH510 pKa = 6.18LISDD514 pKa = 4.5PLNVLGQDD522 pKa = 3.3IQVCYY527 pKa = 10.06NWEE530 pKa = 4.55DD531 pKa = 3.51GTIADD536 pKa = 4.7FDD538 pKa = 5.33DD539 pKa = 3.89IGIVVEE545 pKa = 5.53DD546 pKa = 4.54CDD548 pKa = 4.66CFINIWDD555 pKa = 3.85EE556 pKa = 4.0NGNFLGKK563 pKa = 10.09NPEE566 pKa = 4.04CSEE569 pKa = 4.02VGLFEE574 pKa = 4.43GNYY577 pKa = 9.66YY578 pKa = 10.92GLYY581 pKa = 10.56DD582 pKa = 3.47NYY584 pKa = 10.97RR585 pKa = 11.84PTFCSIMDD593 pKa = 3.67SCSAAGYY600 pKa = 9.62QKK602 pKa = 11.13VNAEE606 pKa = 3.98GFAVGSIHH614 pKa = 6.69NQGFYY619 pKa = 11.1NSDD622 pKa = 3.22SVGFLTDD629 pKa = 3.36STTGEE634 pKa = 4.07YY635 pKa = 10.24TDD637 pKa = 4.28FEE639 pKa = 4.29IVIDD643 pKa = 4.45GEE645 pKa = 4.54LDD647 pKa = 3.25TSKK650 pKa = 11.0LVLKK654 pKa = 9.68WYY656 pKa = 9.98VNGVEE661 pKa = 4.33EE662 pKa = 4.52VGLRR666 pKa = 11.84NQLGAIFPRR675 pKa = 11.84PANNAIEE682 pKa = 4.41IYY684 pKa = 9.26TYY686 pKa = 10.54RR687 pKa = 11.84ITDD690 pKa = 3.58LTGTITAPDD699 pKa = 3.49QVLVYY704 pKa = 10.99DD705 pKa = 4.67DD706 pKa = 4.88FYY708 pKa = 11.71EE709 pKa = 4.57GLLNSDD715 pKa = 4.42FQWXXXXXXXLSTAYY730 pKa = 10.32DD731 pKa = 3.89YY732 pKa = 11.65AWCCNKK738 pKa = 9.94SRR740 pKa = 11.84IDD742 pKa = 3.57PNGEE746 pKa = 3.5II747 pKa = 4.56
MM1 pKa = 7.51KK2 pKa = 10.29SKK4 pKa = 10.89SSLFIAFLFLFSCGGGGGGGSAALPLPIITIAASSLDD41 pKa = 3.82VNIGDD46 pKa = 4.94DD47 pKa = 4.75VDD49 pKa = 4.06ITWSVSNSNSCNATGAWSGIKK70 pKa = 10.19SNSGNEE76 pKa = 4.05TVTVSGAGNNNFGLSCSGEE95 pKa = 4.04GGQASEE101 pKa = 4.46TVTVFAFNLAANNTTLAVDD120 pKa = 3.82EE121 pKa = 5.47DD122 pKa = 4.29GSLINSSVAVQPNTDD137 pKa = 2.99ILVSYY142 pKa = 10.36SLLSSTSNGNLTFNEE157 pKa = 4.39DD158 pKa = 2.91ATINYY163 pKa = 9.29YY164 pKa = 10.0PDD166 pKa = 3.85PNFNGVDD173 pKa = 3.25EE174 pKa = 4.84FTFKK178 pKa = 10.96AIAATRR184 pKa = 11.84SVEE187 pKa = 3.83KK188 pKa = 10.8DD189 pKa = 3.05VTVTINIASINDD201 pKa = 3.57APTIALDD208 pKa = 4.02SEE210 pKa = 5.11SNLSKK215 pKa = 11.0LDD217 pKa = 3.43MVYY220 pKa = 10.72EE221 pKa = 4.18SDD223 pKa = 4.94PIFSFNFADD232 pKa = 3.72VDD234 pKa = 3.88HH235 pKa = 6.95LLEE238 pKa = 5.19DD239 pKa = 3.75LTFTASVDD247 pKa = 3.54GNAVPTTFNQISEE260 pKa = 5.0GYY262 pKa = 10.65GSLQLDD268 pKa = 4.5LSSLGSGGLFDD279 pKa = 4.26LTLSVFDD286 pKa = 4.76GFDD289 pKa = 3.25TATEE293 pKa = 4.12TLNTWFVADD302 pKa = 4.0KK303 pKa = 10.47STVTFDD309 pKa = 4.43QDD311 pKa = 3.93DD312 pKa = 4.09DD313 pKa = 4.88PEE315 pKa = 6.54DD316 pKa = 4.0GVTEE320 pKa = 4.55GSSTSVDD327 pKa = 3.35YY328 pKa = 11.07NVYY331 pKa = 10.24YY332 pKa = 11.02LDD334 pKa = 4.42GNSEE338 pKa = 4.08SRR340 pKa = 11.84GRR342 pKa = 11.84TTYY345 pKa = 11.25LFVADD350 pKa = 4.23SLASEE355 pKa = 4.47EE356 pKa = 4.34DD357 pKa = 3.33RR358 pKa = 11.84ASFRR362 pKa = 11.84SALVRR367 pKa = 11.84SINKK371 pKa = 9.25VKK373 pKa = 10.63EE374 pKa = 3.78SDD376 pKa = 3.1AGEE379 pKa = 4.58FITGFFTIKK388 pKa = 10.16AAEE391 pKa = 3.92PVIPDD396 pKa = 3.91GKK398 pKa = 10.33SPSAIRR404 pKa = 11.84TGCYY408 pKa = 10.39DD409 pKa = 4.32FDD411 pKa = 4.97PNIYY415 pKa = 10.28CIGDD419 pKa = 3.38MDD421 pKa = 4.12TSVFDD426 pKa = 3.46VMYY429 pKa = 9.65PGHH432 pKa = 7.12LLVSTLTMQSGRR444 pKa = 11.84GVNLGNRR451 pKa = 11.84NIQPISSRR459 pKa = 11.84TQNVLMHH466 pKa = 6.52EE467 pKa = 4.86LGHH470 pKa = 6.39AHH472 pKa = 6.91GFMGDD477 pKa = 3.69EE478 pKa = 4.24YY479 pKa = 11.11RR480 pKa = 11.84SDD482 pKa = 3.9DD483 pKa = 4.79DD484 pKa = 5.68RR485 pKa = 11.84DD486 pKa = 3.32VSYY489 pKa = 9.81WADD492 pKa = 3.47LNINTTTQSDD502 pKa = 3.81PALVKK507 pKa = 9.65WKK509 pKa = 10.26HH510 pKa = 6.18LISDD514 pKa = 4.5PLNVLGQDD522 pKa = 3.3IQVCYY527 pKa = 10.06NWEE530 pKa = 4.55DD531 pKa = 3.51GTIADD536 pKa = 4.7FDD538 pKa = 5.33DD539 pKa = 3.89IGIVVEE545 pKa = 5.53DD546 pKa = 4.54CDD548 pKa = 4.66CFINIWDD555 pKa = 3.85EE556 pKa = 4.0NGNFLGKK563 pKa = 10.09NPEE566 pKa = 4.04CSEE569 pKa = 4.02VGLFEE574 pKa = 4.43GNYY577 pKa = 9.66YY578 pKa = 10.92GLYY581 pKa = 10.56DD582 pKa = 3.47NYY584 pKa = 10.97RR585 pKa = 11.84PTFCSIMDD593 pKa = 3.67SCSAAGYY600 pKa = 9.62QKK602 pKa = 11.13VNAEE606 pKa = 3.98GFAVGSIHH614 pKa = 6.69NQGFYY619 pKa = 11.1NSDD622 pKa = 3.22SVGFLTDD629 pKa = 3.36STTGEE634 pKa = 4.07YY635 pKa = 10.24TDD637 pKa = 4.28FEE639 pKa = 4.29IVIDD643 pKa = 4.45GEE645 pKa = 4.54LDD647 pKa = 3.25TSKK650 pKa = 11.0LVLKK654 pKa = 9.68WYY656 pKa = 9.98VNGVEE661 pKa = 4.33EE662 pKa = 4.52VGLRR666 pKa = 11.84NQLGAIFPRR675 pKa = 11.84PANNAIEE682 pKa = 4.41IYY684 pKa = 9.26TYY686 pKa = 10.54RR687 pKa = 11.84ITDD690 pKa = 3.58LTGTITAPDD699 pKa = 3.49QVLVYY704 pKa = 10.99DD705 pKa = 4.67DD706 pKa = 4.88FYY708 pKa = 11.71EE709 pKa = 4.57GLLNSDD715 pKa = 4.42FQWXXXXXXXLSTAYY730 pKa = 10.32DD731 pKa = 3.89YY732 pKa = 11.65AWCCNKK738 pKa = 9.94SRR740 pKa = 11.84IDD742 pKa = 3.57PNGEE746 pKa = 3.5II747 pKa = 4.56
Molecular weight: 80.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J5KLI8|J5KLI8_9GAMM Glucose-inhibited division protein A OS=SAR86 cluster bacterium SAR86B OX=1123867 GN=gidA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.2AGRR28 pKa = 11.84AILANRR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 10.03KK37 pKa = 9.78GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.2AGRR28 pKa = 11.84AILANRR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 10.03KK37 pKa = 9.78GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
523114 |
29 |
1477 |
289.5 |
32.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.17 ± 0.059 | 0.919 ± 0.017 |
6.007 ± 0.045 | 6.54 ± 0.051 |
4.861 ± 0.049 | 6.439 ± 0.057 |
1.692 ± 0.024 | 9.006 ± 0.075 |
7.503 ± 0.074 | 9.345 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.182 ± 0.026 | 6.414 ± 0.056 |
3.47 ± 0.035 | 2.819 ± 0.026 |
3.418 ± 0.04 | 7.594 ± 0.051 |
4.769 ± 0.04 | 5.727 ± 0.053 |
0.998 ± 0.024 | 3.549 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
