
Hordeum vulgare subsp. vulgare (Domesticated barley)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade;
Average proteome isoelectric point is 6.88
Get precalculated fractions of proteins
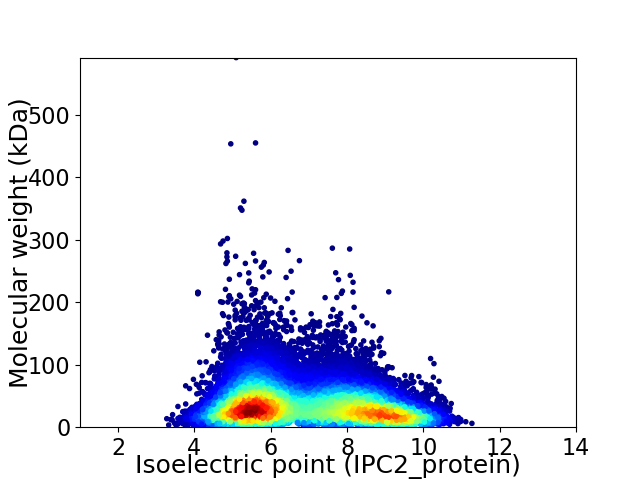
Virtual 2D-PAGE plot for 189799 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M0V9N4|M0V9N4_HORVV Isoform of A0A287PHC5 Uncharacterized protein OS=Hordeum vulgare subsp. vulgare OX=112509 PE=4 SV=1
MM1 pKa = 7.29HH2 pKa = 6.18VVCRR6 pKa = 11.84IFQKK10 pKa = 10.73VGSGPQNGAQYY21 pKa = 9.57GAPYY25 pKa = 9.48MEE27 pKa = 5.18EE28 pKa = 3.88EE29 pKa = 4.76WEE31 pKa = 4.61DD32 pKa = 3.53EE33 pKa = 4.22DD34 pKa = 5.85DD35 pKa = 5.58AIEE38 pKa = 4.09NTPTSGTSTEE48 pKa = 4.18MLAITDD54 pKa = 3.86TASAEE59 pKa = 4.47SNVEE63 pKa = 3.87DD64 pKa = 3.95EE65 pKa = 5.29NIFSGINEE73 pKa = 4.49LVQIQDD79 pKa = 3.35VLIPPEE85 pKa = 4.01IVAPIQAIAPLQAQGLNGTGMGSYY109 pKa = 11.21ADD111 pKa = 4.04GDD113 pKa = 3.9VSLDD117 pKa = 3.98EE118 pKa = 4.6ILQEE122 pKa = 4.05PVSDD126 pKa = 3.64VSVDD130 pKa = 3.41NTGEE134 pKa = 4.11PEE136 pKa = 4.16EE137 pKa = 4.08QSPIDD142 pKa = 3.77DD143 pKa = 4.35HH144 pKa = 7.45FSLADD149 pKa = 4.01LSGCPNQDD157 pKa = 3.0DD158 pKa = 5.21GYY160 pKa = 10.97VRR162 pKa = 11.84QAGLTMWSDD171 pKa = 3.82PSNGDD176 pKa = 3.02QAYY179 pKa = 9.14YY180 pKa = 10.06PIIRR184 pKa = 11.84TYY186 pKa = 11.28GNRR189 pKa = 11.84NHH191 pKa = 6.96ANMALSDD198 pKa = 3.79AEE200 pKa = 4.38YY201 pKa = 10.7FDD203 pKa = 5.01TEE205 pKa = 4.26NDD207 pKa = 3.16TNAYY211 pKa = 9.88FGQQQACPSDD221 pKa = 3.66DD222 pKa = 3.5QNLYY226 pKa = 10.66SGQQQACPSDD236 pKa = 3.67DD237 pKa = 3.54QNLYY241 pKa = 10.79GPSFPQQVGDD251 pKa = 3.53NEE253 pKa = 4.2QFYY256 pKa = 10.32EE257 pKa = 4.49ASSDD261 pKa = 3.79HH262 pKa = 6.38KK263 pKa = 10.72WVDD266 pKa = 4.1GKK268 pKa = 10.75DD269 pKa = 3.2DD270 pKa = 3.87CANVNDD276 pKa = 5.35LSIFDD281 pKa = 4.88DD282 pKa = 6.18DD283 pKa = 5.87IMSLLNASEE292 pKa = 4.64GDD294 pKa = 3.56FSLDD298 pKa = 3.31LLGPVDD304 pKa = 4.16GSNSQFPAASDD315 pKa = 3.86FDD317 pKa = 4.03QKK319 pKa = 11.66DD320 pKa = 3.38EE321 pKa = 4.72AKK323 pKa = 9.97AQYY326 pKa = 9.62GASSSGSRR334 pKa = 11.84EE335 pKa = 3.72NLYY338 pKa = 9.71PDD340 pKa = 4.64SMVTDD345 pKa = 4.67LPMDD349 pKa = 4.64DD350 pKa = 4.46NVGKK354 pKa = 10.83RR355 pKa = 11.84FGKK358 pKa = 8.89FANMLGSYY366 pKa = 7.88PAPPAMASEE375 pKa = 4.84FPPTTGKK382 pKa = 10.59SIAALSGPSQIRR394 pKa = 11.84VTAGIVQLGDD404 pKa = 3.2HH405 pKa = 6.59SFTGNSDD412 pKa = 2.93NWPLQKK418 pKa = 10.88NGVFNLLLSFTVEE431 pKa = 4.5SNVSTKK437 pKa = 10.88LIGFDD442 pKa = 4.87DD443 pKa = 4.69EE444 pKa = 4.57PATTRR449 pKa = 11.84VSTVPTVLRR458 pKa = 11.84GGLYY462 pKa = 10.61LFFVSAMILMLSFKK476 pKa = 10.49VGSCIYY482 pKa = 10.71SRR484 pKa = 3.98
MM1 pKa = 7.29HH2 pKa = 6.18VVCRR6 pKa = 11.84IFQKK10 pKa = 10.73VGSGPQNGAQYY21 pKa = 9.57GAPYY25 pKa = 9.48MEE27 pKa = 5.18EE28 pKa = 3.88EE29 pKa = 4.76WEE31 pKa = 4.61DD32 pKa = 3.53EE33 pKa = 4.22DD34 pKa = 5.85DD35 pKa = 5.58AIEE38 pKa = 4.09NTPTSGTSTEE48 pKa = 4.18MLAITDD54 pKa = 3.86TASAEE59 pKa = 4.47SNVEE63 pKa = 3.87DD64 pKa = 3.95EE65 pKa = 5.29NIFSGINEE73 pKa = 4.49LVQIQDD79 pKa = 3.35VLIPPEE85 pKa = 4.01IVAPIQAIAPLQAQGLNGTGMGSYY109 pKa = 11.21ADD111 pKa = 4.04GDD113 pKa = 3.9VSLDD117 pKa = 3.98EE118 pKa = 4.6ILQEE122 pKa = 4.05PVSDD126 pKa = 3.64VSVDD130 pKa = 3.41NTGEE134 pKa = 4.11PEE136 pKa = 4.16EE137 pKa = 4.08QSPIDD142 pKa = 3.77DD143 pKa = 4.35HH144 pKa = 7.45FSLADD149 pKa = 4.01LSGCPNQDD157 pKa = 3.0DD158 pKa = 5.21GYY160 pKa = 10.97VRR162 pKa = 11.84QAGLTMWSDD171 pKa = 3.82PSNGDD176 pKa = 3.02QAYY179 pKa = 9.14YY180 pKa = 10.06PIIRR184 pKa = 11.84TYY186 pKa = 11.28GNRR189 pKa = 11.84NHH191 pKa = 6.96ANMALSDD198 pKa = 3.79AEE200 pKa = 4.38YY201 pKa = 10.7FDD203 pKa = 5.01TEE205 pKa = 4.26NDD207 pKa = 3.16TNAYY211 pKa = 9.88FGQQQACPSDD221 pKa = 3.66DD222 pKa = 3.5QNLYY226 pKa = 10.66SGQQQACPSDD236 pKa = 3.67DD237 pKa = 3.54QNLYY241 pKa = 10.79GPSFPQQVGDD251 pKa = 3.53NEE253 pKa = 4.2QFYY256 pKa = 10.32EE257 pKa = 4.49ASSDD261 pKa = 3.79HH262 pKa = 6.38KK263 pKa = 10.72WVDD266 pKa = 4.1GKK268 pKa = 10.75DD269 pKa = 3.2DD270 pKa = 3.87CANVNDD276 pKa = 5.35LSIFDD281 pKa = 4.88DD282 pKa = 6.18DD283 pKa = 5.87IMSLLNASEE292 pKa = 4.64GDD294 pKa = 3.56FSLDD298 pKa = 3.31LLGPVDD304 pKa = 4.16GSNSQFPAASDD315 pKa = 3.86FDD317 pKa = 4.03QKK319 pKa = 11.66DD320 pKa = 3.38EE321 pKa = 4.72AKK323 pKa = 9.97AQYY326 pKa = 9.62GASSSGSRR334 pKa = 11.84EE335 pKa = 3.72NLYY338 pKa = 9.71PDD340 pKa = 4.64SMVTDD345 pKa = 4.67LPMDD349 pKa = 4.64DD350 pKa = 4.46NVGKK354 pKa = 10.83RR355 pKa = 11.84FGKK358 pKa = 8.89FANMLGSYY366 pKa = 7.88PAPPAMASEE375 pKa = 4.84FPPTTGKK382 pKa = 10.59SIAALSGPSQIRR394 pKa = 11.84VTAGIVQLGDD404 pKa = 3.2HH405 pKa = 6.59SFTGNSDD412 pKa = 2.93NWPLQKK418 pKa = 10.88NGVFNLLLSFTVEE431 pKa = 4.5SNVSTKK437 pKa = 10.88LIGFDD442 pKa = 4.87DD443 pKa = 4.69EE444 pKa = 4.57PATTRR449 pKa = 11.84VSTVPTVLRR458 pKa = 11.84GGLYY462 pKa = 10.61LFFVSAMILMLSFKK476 pKa = 10.49VGSCIYY482 pKa = 10.71SRR484 pKa = 3.98
Molecular weight: 52.3 kDa
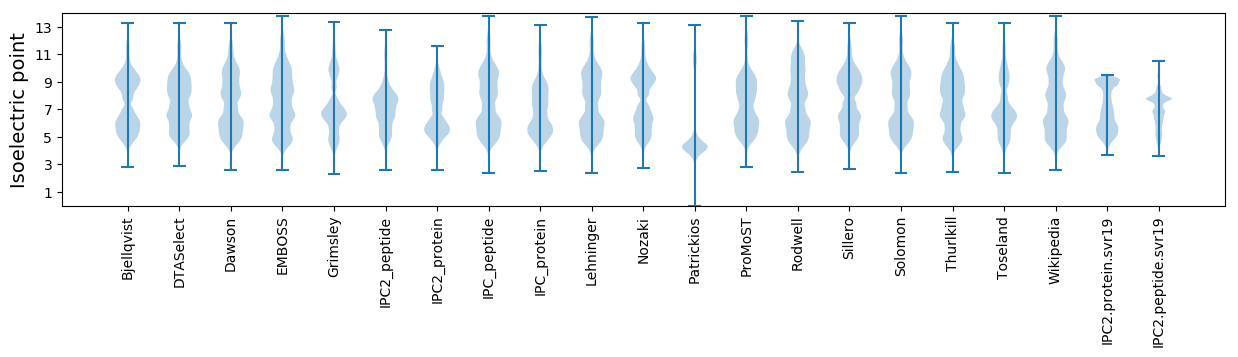
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A287JF95|A0A287JF95_HORVV Isoform of A0A287JEV7 Mediator of RNA polymerase II transcription subunit 13 OS=Hordeum vulgare subsp. vulgare OX=112509 PE=3 SV=1
PP1 pKa = 7.93PIPTPQRR8 pKa = 11.84HH9 pKa = 5.15RR10 pKa = 11.84PPWLSLTPASTPRR23 pKa = 11.84APPLSAPPPPPLAPPRR39 pKa = 11.84VPRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84PPSPPPPSPALTSPPRR62 pKa = 11.84APRR65 pKa = 11.84PVAPRR70 pKa = 11.84PSAPPWAPRR79 pKa = 11.84HH80 pKa = 5.45PRR82 pKa = 11.84ISSGPPPAA90 pKa = 5.69
PP1 pKa = 7.93PIPTPQRR8 pKa = 11.84HH9 pKa = 5.15RR10 pKa = 11.84PPWLSLTPASTPRR23 pKa = 11.84APPLSAPPPPPLAPPRR39 pKa = 11.84VPRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84PPSPPPPSPALTSPPRR62 pKa = 11.84APRR65 pKa = 11.84PVAPRR70 pKa = 11.84PSAPPWAPRR79 pKa = 11.84HH80 pKa = 5.45PRR82 pKa = 11.84ISSGPPPAA90 pKa = 5.69
Molecular weight: 9.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
68226991 |
29 |
5383 |
359.5 |
39.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.23 ± 0.007 | 1.932 ± 0.003 |
5.36 ± 0.004 | 6.006 ± 0.007 |
3.787 ± 0.004 | 6.943 ± 0.006 |
2.554 ± 0.003 | 4.71 ± 0.005 |
5.188 ± 0.005 | 9.711 ± 0.007 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.44 ± 0.003 | 3.683 ± 0.004 |
5.366 ± 0.006 | 3.68 ± 0.004 |
6.04 ± 0.006 | 8.483 ± 0.006 |
4.896 ± 0.003 | 6.687 ± 0.004 |
1.262 ± 0.002 | 2.682 ± 0.003 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
