
Triticum aestivum (Wheat)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae;
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
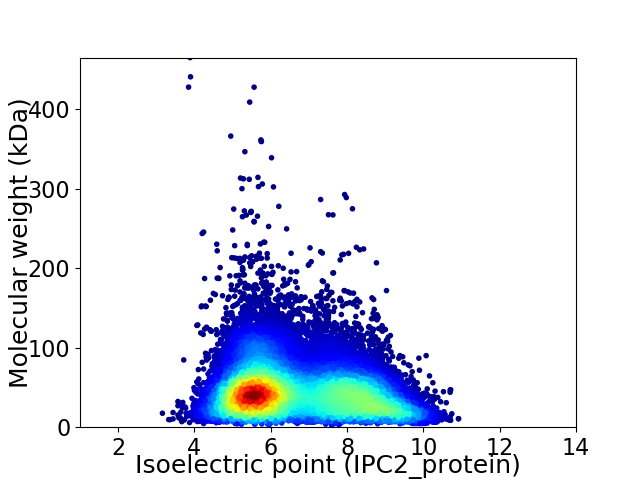
Virtual 2D-PAGE plot for 130673 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
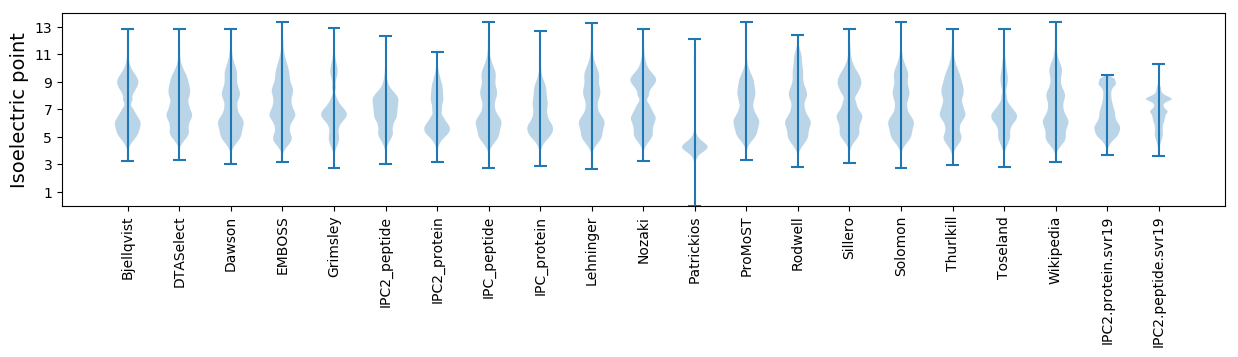
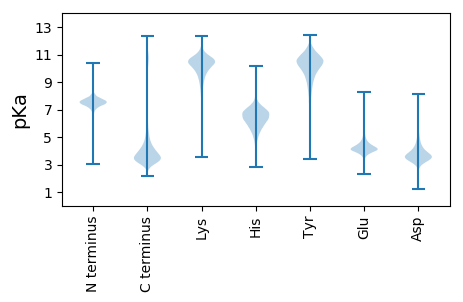
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3B6ARP4|A0A3B6ARP4_WHEAT Uncharacterized protein OS=Triticum aestivum OX=4565 PE=4 SV=1
MM1 pKa = 7.17GKK3 pKa = 10.26AYY5 pKa = 10.28RR6 pKa = 11.84KK7 pKa = 9.97KK8 pKa = 10.87LGVNKK13 pKa = 10.13AATPAGGKK21 pKa = 10.19GKK23 pKa = 10.49GAGGGNAKK31 pKa = 10.23RR32 pKa = 11.84SGGAAGAGGAGPGGGAKK49 pKa = 10.22GAGGAGWGGASTFSATYY66 pKa = 10.04HH67 pKa = 5.68STFQTAVNVTNEE79 pKa = 4.49YY80 pKa = 9.74YY81 pKa = 10.53NQWGNDD87 pKa = 3.44CSSDD91 pKa = 4.14DD92 pKa = 4.92DD93 pKa = 4.9GPPSAPADD101 pKa = 3.64DD102 pKa = 4.42STFEE106 pKa = 4.09TAVNDD111 pKa = 3.7TNEE114 pKa = 4.12YY115 pKa = 8.71YY116 pKa = 10.09QEE118 pKa = 4.49EE119 pKa = 4.32EE120 pKa = 4.83DD121 pKa = 4.48NNCSSPFAPADD132 pKa = 3.21HH133 pKa = 6.51STFEE137 pKa = 4.2TAVKK141 pKa = 10.2IMNEE145 pKa = 4.09HH146 pKa = 6.47YY147 pKa = 10.99DD148 pKa = 3.57DD149 pKa = 5.79DD150 pKa = 4.87EE151 pKa = 5.0NLGLSMDD158 pKa = 3.95SEE160 pKa = 4.72EE161 pKa = 5.36EE162 pKa = 4.38GVQDD166 pKa = 4.93AEE168 pKa = 4.44PVSDD172 pKa = 5.09ADD174 pKa = 5.29GEE176 pKa = 4.4GDD178 pKa = 4.18DD179 pKa = 5.42DD180 pKa = 5.36AGSGDD185 pKa = 4.1DD186 pKa = 5.13HH187 pKa = 8.31FGDD190 pKa = 4.96DD191 pKa = 5.5DD192 pKa = 5.48YY193 pKa = 11.97YY194 pKa = 11.61DD195 pKa = 5.53DD196 pKa = 6.76DD197 pKa = 4.42IAGCFDD203 pKa = 3.62AFYY206 pKa = 10.92DD207 pKa = 4.59PGDD210 pKa = 4.28AGDD213 pKa = 5.56GADD216 pKa = 4.3DD217 pKa = 3.21WWW219 pKa = 5.75
MM1 pKa = 7.17GKK3 pKa = 10.26AYY5 pKa = 10.28RR6 pKa = 11.84KK7 pKa = 9.97KK8 pKa = 10.87LGVNKK13 pKa = 10.13AATPAGGKK21 pKa = 10.19GKK23 pKa = 10.49GAGGGNAKK31 pKa = 10.23RR32 pKa = 11.84SGGAAGAGGAGPGGGAKK49 pKa = 10.22GAGGAGWGGASTFSATYY66 pKa = 10.04HH67 pKa = 5.68STFQTAVNVTNEE79 pKa = 4.49YY80 pKa = 9.74YY81 pKa = 10.53NQWGNDD87 pKa = 3.44CSSDD91 pKa = 4.14DD92 pKa = 4.92DD93 pKa = 4.9GPPSAPADD101 pKa = 3.64DD102 pKa = 4.42STFEE106 pKa = 4.09TAVNDD111 pKa = 3.7TNEE114 pKa = 4.12YY115 pKa = 8.71YY116 pKa = 10.09QEE118 pKa = 4.49EE119 pKa = 4.32EE120 pKa = 4.83DD121 pKa = 4.48NNCSSPFAPADD132 pKa = 3.21HH133 pKa = 6.51STFEE137 pKa = 4.2TAVKK141 pKa = 10.2IMNEE145 pKa = 4.09HH146 pKa = 6.47YY147 pKa = 10.99DD148 pKa = 3.57DD149 pKa = 5.79DD150 pKa = 4.87EE151 pKa = 5.0NLGLSMDD158 pKa = 3.95SEE160 pKa = 4.72EE161 pKa = 5.36EE162 pKa = 4.38GVQDD166 pKa = 4.93AEE168 pKa = 4.44PVSDD172 pKa = 5.09ADD174 pKa = 5.29GEE176 pKa = 4.4GDD178 pKa = 4.18DD179 pKa = 5.42DD180 pKa = 5.36AGSGDD185 pKa = 4.1DD186 pKa = 5.13HH187 pKa = 8.31FGDD190 pKa = 4.96DD191 pKa = 5.5DD192 pKa = 5.48YY193 pKa = 11.97YY194 pKa = 11.61DD195 pKa = 5.53DD196 pKa = 6.76DD197 pKa = 4.42IAGCFDD203 pKa = 3.62AFYY206 pKa = 10.92DD207 pKa = 4.59PGDD210 pKa = 4.28AGDD213 pKa = 5.56GADD216 pKa = 4.3DD217 pKa = 3.21WWW219 pKa = 5.75
Molecular weight: 22.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3B6QD59|A0A3B6QD59_WHEAT Uncharacterized protein OS=Triticum aestivum OX=4565 PE=4 SV=1
MM1 pKa = 7.07YY2 pKa = 10.41LVYY5 pKa = 10.35YY6 pKa = 7.86YY7 pKa = 9.57QHH9 pKa = 7.08IIKK12 pKa = 10.1SCLISTTPLLLLAKK26 pKa = 9.32VHH28 pKa = 6.21IFVLLRR34 pKa = 11.84FSPMHH39 pKa = 5.88IHH41 pKa = 6.17RR42 pKa = 11.84TLPSPCVSSVAFPPHH57 pKa = 6.26SKK59 pKa = 10.13KK60 pKa = 10.6ASRR63 pKa = 11.84PSTAAAHH70 pKa = 5.74HH71 pKa = 6.08RR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84HH76 pKa = 4.64GRR78 pKa = 11.84GHH80 pKa = 6.27GAGHH84 pKa = 7.02GLLLPPAARR93 pKa = 11.84LGRR96 pKa = 11.84HH97 pKa = 6.25LAPPPPPLPRR107 pKa = 11.84RR108 pKa = 11.84APPGPRR114 pKa = 11.84TRR116 pKa = 11.84RR117 pKa = 11.84ARR119 pKa = 11.84PLRR122 pKa = 11.84RR123 pKa = 11.84GRR125 pKa = 11.84PRR127 pKa = 11.84GRR129 pKa = 11.84RR130 pKa = 11.84AGGGTASSAAAGAAPSGGVGEE151 pKa = 4.38AQGAGAPAAGVAGGGGADD169 pKa = 3.12GRR171 pKa = 11.84VRR173 pKa = 11.84RR174 pKa = 11.84GGRR177 pKa = 11.84GGPRR181 pKa = 11.84RR182 pKa = 11.84LRR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84RR187 pKa = 11.84VPVQDD192 pKa = 3.31PRR194 pKa = 11.84VGRR197 pKa = 11.84RR198 pKa = 11.84APGRR202 pKa = 11.84RR203 pKa = 11.84EE204 pKa = 3.51GAPLVRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84RR213 pKa = 11.84APARR217 pKa = 11.84RR218 pKa = 11.84VPARR222 pKa = 11.84ARR224 pKa = 11.84PAPPRR229 pKa = 11.84RR230 pKa = 11.84PPPAPRR236 pKa = 11.84RR237 pKa = 11.84PPRR240 pKa = 11.84LLRR243 pKa = 11.84RR244 pKa = 11.84PRR246 pKa = 11.84GGRR249 pKa = 11.84RR250 pKa = 11.84AGAGVHH256 pKa = 6.58AGRR259 pKa = 11.84HH260 pKa = 5.27AGGPSPPRR268 pKa = 11.84RR269 pKa = 11.84HH270 pKa = 5.94AAAHH274 pKa = 6.67LAPPHH279 pKa = 6.5AGPPRR284 pKa = 11.84RR285 pKa = 11.84GRR287 pKa = 11.84RR288 pKa = 11.84AGAPPRR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84LHH298 pKa = 5.56RR299 pKa = 11.84ARR301 pKa = 11.84RR302 pKa = 11.84RR303 pKa = 11.84VGLQRR308 pKa = 11.84APRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84RR314 pKa = 11.84PRR316 pKa = 11.84AALRR320 pKa = 11.84PGVGLRR326 pKa = 11.84GHH328 pKa = 7.31LLGGRR333 pKa = 11.84GPRR336 pKa = 11.84PGRR339 pKa = 11.84GGRR342 pKa = 11.84RR343 pKa = 11.84LAGLRR348 pKa = 11.84RR349 pKa = 11.84PLLPPHH355 pKa = 6.15RR356 pKa = 11.84HH357 pKa = 5.08RR358 pKa = 11.84VQEE361 pKa = 3.85VRR363 pKa = 11.84RR364 pKa = 11.84VRR366 pKa = 11.84LRR368 pKa = 11.84RR369 pKa = 11.84AAPRR373 pKa = 11.84GPHH376 pKa = 6.08RR377 pKa = 11.84LAGRR381 pKa = 11.84RR382 pKa = 11.84RR383 pKa = 11.84AGPGRR388 pKa = 11.84RR389 pKa = 11.84RR390 pKa = 11.84RR391 pKa = 11.84PVPRR395 pKa = 11.84GQGAAARR402 pKa = 11.84EE403 pKa = 4.21GRR405 pKa = 11.84RR406 pKa = 11.84GGGARR411 pKa = 11.84GRR413 pKa = 11.84QAGRR417 pKa = 11.84RR418 pKa = 11.84LL419 pKa = 3.31
MM1 pKa = 7.07YY2 pKa = 10.41LVYY5 pKa = 10.35YY6 pKa = 7.86YY7 pKa = 9.57QHH9 pKa = 7.08IIKK12 pKa = 10.1SCLISTTPLLLLAKK26 pKa = 9.32VHH28 pKa = 6.21IFVLLRR34 pKa = 11.84FSPMHH39 pKa = 5.88IHH41 pKa = 6.17RR42 pKa = 11.84TLPSPCVSSVAFPPHH57 pKa = 6.26SKK59 pKa = 10.13KK60 pKa = 10.6ASRR63 pKa = 11.84PSTAAAHH70 pKa = 5.74HH71 pKa = 6.08RR72 pKa = 11.84RR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84HH76 pKa = 4.64GRR78 pKa = 11.84GHH80 pKa = 6.27GAGHH84 pKa = 7.02GLLLPPAARR93 pKa = 11.84LGRR96 pKa = 11.84HH97 pKa = 6.25LAPPPPPLPRR107 pKa = 11.84RR108 pKa = 11.84APPGPRR114 pKa = 11.84TRR116 pKa = 11.84RR117 pKa = 11.84ARR119 pKa = 11.84PLRR122 pKa = 11.84RR123 pKa = 11.84GRR125 pKa = 11.84PRR127 pKa = 11.84GRR129 pKa = 11.84RR130 pKa = 11.84AGGGTASSAAAGAAPSGGVGEE151 pKa = 4.38AQGAGAPAAGVAGGGGADD169 pKa = 3.12GRR171 pKa = 11.84VRR173 pKa = 11.84RR174 pKa = 11.84GGRR177 pKa = 11.84GGPRR181 pKa = 11.84RR182 pKa = 11.84LRR184 pKa = 11.84RR185 pKa = 11.84RR186 pKa = 11.84RR187 pKa = 11.84VPVQDD192 pKa = 3.31PRR194 pKa = 11.84VGRR197 pKa = 11.84RR198 pKa = 11.84APGRR202 pKa = 11.84RR203 pKa = 11.84EE204 pKa = 3.51GAPLVRR210 pKa = 11.84RR211 pKa = 11.84RR212 pKa = 11.84RR213 pKa = 11.84APARR217 pKa = 11.84RR218 pKa = 11.84VPARR222 pKa = 11.84ARR224 pKa = 11.84PAPPRR229 pKa = 11.84RR230 pKa = 11.84PPPAPRR236 pKa = 11.84RR237 pKa = 11.84PPRR240 pKa = 11.84LLRR243 pKa = 11.84RR244 pKa = 11.84PRR246 pKa = 11.84GGRR249 pKa = 11.84RR250 pKa = 11.84AGAGVHH256 pKa = 6.58AGRR259 pKa = 11.84HH260 pKa = 5.27AGGPSPPRR268 pKa = 11.84RR269 pKa = 11.84HH270 pKa = 5.94AAAHH274 pKa = 6.67LAPPHH279 pKa = 6.5AGPPRR284 pKa = 11.84RR285 pKa = 11.84GRR287 pKa = 11.84RR288 pKa = 11.84AGAPPRR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84LHH298 pKa = 5.56RR299 pKa = 11.84ARR301 pKa = 11.84RR302 pKa = 11.84RR303 pKa = 11.84VGLQRR308 pKa = 11.84APRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84RR314 pKa = 11.84PRR316 pKa = 11.84AALRR320 pKa = 11.84PGVGLRR326 pKa = 11.84GHH328 pKa = 7.31LLGGRR333 pKa = 11.84GPRR336 pKa = 11.84PGRR339 pKa = 11.84GGRR342 pKa = 11.84RR343 pKa = 11.84LAGLRR348 pKa = 11.84RR349 pKa = 11.84PLLPPHH355 pKa = 6.15RR356 pKa = 11.84HH357 pKa = 5.08RR358 pKa = 11.84VQEE361 pKa = 3.85VRR363 pKa = 11.84RR364 pKa = 11.84VRR366 pKa = 11.84LRR368 pKa = 11.84RR369 pKa = 11.84AAPRR373 pKa = 11.84GPHH376 pKa = 6.08RR377 pKa = 11.84LAGRR381 pKa = 11.84RR382 pKa = 11.84RR383 pKa = 11.84AGPGRR388 pKa = 11.84RR389 pKa = 11.84RR390 pKa = 11.84RR391 pKa = 11.84PVPRR395 pKa = 11.84GQGAAARR402 pKa = 11.84EE403 pKa = 4.21GRR405 pKa = 11.84RR406 pKa = 11.84GGGARR411 pKa = 11.84GRR413 pKa = 11.84QAGRR417 pKa = 11.84RR418 pKa = 11.84LL419 pKa = 3.31
Molecular weight: 45.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
58552452 |
15 |
5359 |
448.1 |
49.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.805 ± 0.009 | 1.937 ± 0.003 |
5.533 ± 0.005 | 6.069 ± 0.007 |
3.698 ± 0.004 | 7.165 ± 0.006 |
2.51 ± 0.003 | 4.462 ± 0.005 |
5.065 ± 0.006 | 9.714 ± 0.008 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.501 ± 0.003 | 3.593 ± 0.004 |
5.459 ± 0.006 | 3.553 ± 0.005 |
6.015 ± 0.006 | 8.301 ± 0.007 |
4.852 ± 0.004 | 6.817 ± 0.005 |
1.288 ± 0.002 | 2.634 ± 0.003 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
