
Pseudo-nitzschia multistriata
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Ochrophyta; Bacillariophyta; Bacillariophyceae; Bacillariophycidae; Bacillariales; Bacillariaceae; Pseudo-nitzschia
Average proteome isoelectric point is 6.35
Get precalculated fractions of proteins
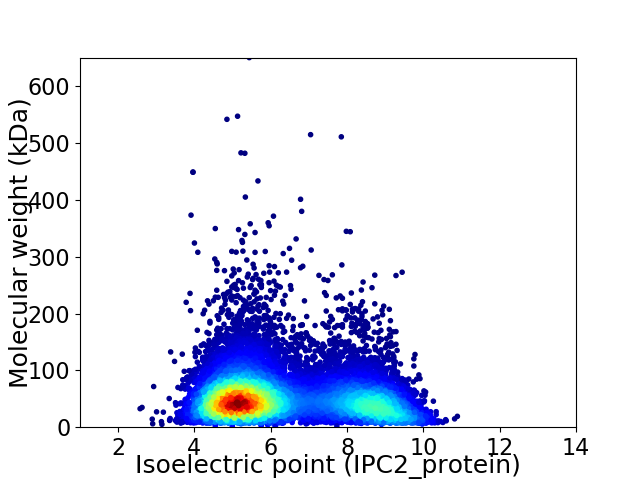
Virtual 2D-PAGE plot for 11907 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A448ZGN2|A0A448ZGN2_9STRA Helicase ATP-binding domain-containing protein OS=Pseudo-nitzschia multistriata OX=183589 GN=PSNMU_V1.4_AUG-EV-PASAV3_0081690 PE=4 SV=1
MM1 pKa = 7.53KK2 pKa = 7.78PTRR5 pKa = 11.84SSRR8 pKa = 11.84FLLLVLLFLVSFLVRR23 pKa = 11.84STMGEE28 pKa = 4.03TLLVKK33 pKa = 10.61AQDD36 pKa = 3.96EE37 pKa = 4.67DD38 pKa = 3.88EE39 pKa = 4.98DD40 pKa = 4.61AADD43 pKa = 4.1TPAVADD49 pKa = 4.22DD50 pKa = 5.01GGDD53 pKa = 3.42EE54 pKa = 4.5TEE56 pKa = 4.87TDD58 pKa = 3.71TEE60 pKa = 4.12TDD62 pKa = 3.34PAAEE66 pKa = 4.53GDD68 pKa = 3.9AEE70 pKa = 4.52GDD72 pKa = 3.6GDD74 pKa = 5.35DD75 pKa = 4.59GDD77 pKa = 5.06GEE79 pKa = 4.28EE80 pKa = 5.47GDD82 pKa = 5.26DD83 pKa = 4.28GDD85 pKa = 4.62EE86 pKa = 4.48EE87 pKa = 4.96SDD89 pKa = 3.49EE90 pKa = 4.3TGEE93 pKa = 4.46EE94 pKa = 4.2GDD96 pKa = 5.68AEE98 pKa = 5.0DD99 pKa = 5.34DD100 pKa = 3.98DD101 pKa = 5.89EE102 pKa = 5.15GFQVEE107 pKa = 4.37DD108 pKa = 4.17LLPGFEE114 pKa = 4.71NTDD117 pKa = 2.57ADD119 pKa = 4.12NFVKK123 pKa = 10.56GYY125 pKa = 10.38ILGNPEE131 pKa = 4.37YY132 pKa = 10.28YY133 pKa = 9.83QEE135 pKa = 5.47LYY137 pKa = 10.36VAPKK141 pKa = 10.62AVTKK145 pKa = 10.22FNNLPRR151 pKa = 11.84FGHH154 pKa = 6.18SDD156 pKa = 3.69DD157 pKa = 4.8FSLLMGDD164 pKa = 4.43YY165 pKa = 9.83PDD167 pKa = 4.63HH168 pKa = 7.05YY169 pKa = 11.24YY170 pKa = 11.07LGLLFVGTLILTIFFFWGIAIGIFKK195 pKa = 10.61CIGPKK200 pKa = 10.06HH201 pKa = 6.09VGFLAGFPFQVEE213 pKa = 4.68GKK215 pKa = 9.31KK216 pKa = 10.07SAAGRR221 pKa = 11.84VVFSVGAWMMIVSTIFLISLGLPRR245 pKa = 11.84MQYY248 pKa = 10.91LSDD251 pKa = 4.45SIAFTADD258 pKa = 3.5EE259 pKa = 3.96IQIVEE264 pKa = 4.44KK265 pKa = 9.45EE266 pKa = 3.97VKK268 pKa = 10.75YY269 pKa = 10.7IVTTLLVVSAKK280 pKa = 9.08TKK282 pKa = 9.75PIRR285 pKa = 11.84DD286 pKa = 3.76EE287 pKa = 4.54LVDD290 pKa = 3.8FLKK293 pKa = 10.62RR294 pKa = 11.84DD295 pKa = 3.49ICPLEE300 pKa = 4.57PGSDD304 pKa = 3.3TEE306 pKa = 4.22DD307 pKa = 3.18QVRR310 pKa = 11.84SVGEE314 pKa = 3.9DD315 pKa = 3.07TYY317 pKa = 11.93DD318 pKa = 3.94AMTDD322 pKa = 3.33LEE324 pKa = 5.06DD325 pKa = 5.01FIEE328 pKa = 5.88GYY330 pKa = 10.82LLDD333 pKa = 4.6VEE335 pKa = 5.82DD336 pKa = 5.74GVNTTNTFTSNILAVTDD353 pKa = 3.41MAQFVGNPVVTAIVFPYY370 pKa = 9.57FIIPIILLVAVGMGWYY386 pKa = 9.91DD387 pKa = 3.97FYY389 pKa = 11.89SEE391 pKa = 4.22TFYY394 pKa = 11.68SFVTWLVVPLLVLLTILAFVTATTLVVAIQANADD428 pKa = 3.36ICVSSGAPEE437 pKa = 4.16DD438 pKa = 4.35SILGVLNNLDD448 pKa = 4.56LEE450 pKa = 4.9DD451 pKa = 5.57DD452 pKa = 4.26SGSQSKK458 pKa = 10.81DD459 pKa = 3.28DD460 pKa = 4.91NGPSDD465 pKa = 3.52NFFYY469 pKa = 10.93DD470 pKa = 2.72IMVFYY475 pKa = 8.94THH477 pKa = 6.47QCTKK481 pKa = 9.45PNPWEE486 pKa = 4.0FMEE489 pKa = 4.5GHH491 pKa = 6.0YY492 pKa = 11.08ADD494 pKa = 4.74LARR497 pKa = 11.84GRR499 pKa = 11.84NILGEE504 pKa = 4.43FIRR507 pKa = 11.84SIEE510 pKa = 4.06DD511 pKa = 3.3TTLLEE516 pKa = 4.43LSQEE520 pKa = 4.48CGMEE524 pKa = 3.8YY525 pKa = 11.05GPIVVLLKK533 pKa = 10.68SLQDD537 pKa = 4.59LITILSQTSIRR548 pKa = 11.84ALTLMSCRR556 pKa = 11.84NVVPLYY562 pKa = 8.76TAWIYY567 pKa = 11.17EE568 pKa = 4.57STCTLAPVSLNLVFMCIVCIAFFGMVCITFRR599 pKa = 11.84GAYY602 pKa = 9.35YY603 pKa = 10.03PIDD606 pKa = 4.19FYY608 pKa = 11.76YY609 pKa = 11.06YY610 pKa = 10.35DD611 pKa = 3.3ATGRR615 pKa = 11.84GDD617 pKa = 3.52KK618 pKa = 10.69QEE620 pKa = 5.08LYY622 pKa = 8.74PTDD625 pKa = 3.58EE626 pKa = 4.52VGDD629 pKa = 3.97SGLADD634 pKa = 3.74TPCNEE639 pKa = 4.27KK640 pKa = 10.69VLEE643 pKa = 4.16EE644 pKa = 4.99QEE646 pKa = 4.36DD647 pKa = 3.75QCIDD651 pKa = 3.01ATEE654 pKa = 3.96AMEE657 pKa = 4.2EE658 pKa = 4.19SIEE661 pKa = 3.87IQIYY665 pKa = 10.33HH666 pKa = 7.27DD667 pKa = 4.51DD668 pKa = 3.8EE669 pKa = 4.5EE670 pKa = 5.75KK671 pKa = 10.66EE672 pKa = 4.15YY673 pKa = 9.86EE674 pKa = 4.18TDD676 pKa = 5.1DD677 pKa = 5.65DD678 pKa = 4.36EE679 pKa = 5.51QDD681 pKa = 3.3HH682 pKa = 6.55EE683 pKa = 5.72KK684 pKa = 11.21NFLLDD689 pKa = 3.65KK690 pKa = 10.67YY691 pKa = 9.36GEE693 pKa = 4.19PMEE696 pKa = 5.25GPDD699 pKa = 3.83AAQHH703 pKa = 5.65SVGTNGSYY711 pKa = 10.33RR712 pKa = 11.84GIDD715 pKa = 3.31RR716 pKa = 11.84NGDD719 pKa = 3.17SHH721 pKa = 7.78AVYY724 pKa = 10.74SDD726 pKa = 4.85DD727 pKa = 4.02DD728 pKa = 4.52DD729 pKa = 5.17EE730 pKa = 4.74MTFHH734 pKa = 6.58STFDD738 pKa = 3.91DD739 pKa = 4.17EE740 pKa = 5.16YY741 pKa = 9.5EE742 pKa = 4.24YY743 pKa = 11.69NSAAEE748 pKa = 4.32EE749 pKa = 3.97EE750 pKa = 4.95SEE752 pKa = 4.33TFSYY756 pKa = 11.28VSS758 pKa = 3.3
MM1 pKa = 7.53KK2 pKa = 7.78PTRR5 pKa = 11.84SSRR8 pKa = 11.84FLLLVLLFLVSFLVRR23 pKa = 11.84STMGEE28 pKa = 4.03TLLVKK33 pKa = 10.61AQDD36 pKa = 3.96EE37 pKa = 4.67DD38 pKa = 3.88EE39 pKa = 4.98DD40 pKa = 4.61AADD43 pKa = 4.1TPAVADD49 pKa = 4.22DD50 pKa = 5.01GGDD53 pKa = 3.42EE54 pKa = 4.5TEE56 pKa = 4.87TDD58 pKa = 3.71TEE60 pKa = 4.12TDD62 pKa = 3.34PAAEE66 pKa = 4.53GDD68 pKa = 3.9AEE70 pKa = 4.52GDD72 pKa = 3.6GDD74 pKa = 5.35DD75 pKa = 4.59GDD77 pKa = 5.06GEE79 pKa = 4.28EE80 pKa = 5.47GDD82 pKa = 5.26DD83 pKa = 4.28GDD85 pKa = 4.62EE86 pKa = 4.48EE87 pKa = 4.96SDD89 pKa = 3.49EE90 pKa = 4.3TGEE93 pKa = 4.46EE94 pKa = 4.2GDD96 pKa = 5.68AEE98 pKa = 5.0DD99 pKa = 5.34DD100 pKa = 3.98DD101 pKa = 5.89EE102 pKa = 5.15GFQVEE107 pKa = 4.37DD108 pKa = 4.17LLPGFEE114 pKa = 4.71NTDD117 pKa = 2.57ADD119 pKa = 4.12NFVKK123 pKa = 10.56GYY125 pKa = 10.38ILGNPEE131 pKa = 4.37YY132 pKa = 10.28YY133 pKa = 9.83QEE135 pKa = 5.47LYY137 pKa = 10.36VAPKK141 pKa = 10.62AVTKK145 pKa = 10.22FNNLPRR151 pKa = 11.84FGHH154 pKa = 6.18SDD156 pKa = 3.69DD157 pKa = 4.8FSLLMGDD164 pKa = 4.43YY165 pKa = 9.83PDD167 pKa = 4.63HH168 pKa = 7.05YY169 pKa = 11.24YY170 pKa = 11.07LGLLFVGTLILTIFFFWGIAIGIFKK195 pKa = 10.61CIGPKK200 pKa = 10.06HH201 pKa = 6.09VGFLAGFPFQVEE213 pKa = 4.68GKK215 pKa = 9.31KK216 pKa = 10.07SAAGRR221 pKa = 11.84VVFSVGAWMMIVSTIFLISLGLPRR245 pKa = 11.84MQYY248 pKa = 10.91LSDD251 pKa = 4.45SIAFTADD258 pKa = 3.5EE259 pKa = 3.96IQIVEE264 pKa = 4.44KK265 pKa = 9.45EE266 pKa = 3.97VKK268 pKa = 10.75YY269 pKa = 10.7IVTTLLVVSAKK280 pKa = 9.08TKK282 pKa = 9.75PIRR285 pKa = 11.84DD286 pKa = 3.76EE287 pKa = 4.54LVDD290 pKa = 3.8FLKK293 pKa = 10.62RR294 pKa = 11.84DD295 pKa = 3.49ICPLEE300 pKa = 4.57PGSDD304 pKa = 3.3TEE306 pKa = 4.22DD307 pKa = 3.18QVRR310 pKa = 11.84SVGEE314 pKa = 3.9DD315 pKa = 3.07TYY317 pKa = 11.93DD318 pKa = 3.94AMTDD322 pKa = 3.33LEE324 pKa = 5.06DD325 pKa = 5.01FIEE328 pKa = 5.88GYY330 pKa = 10.82LLDD333 pKa = 4.6VEE335 pKa = 5.82DD336 pKa = 5.74GVNTTNTFTSNILAVTDD353 pKa = 3.41MAQFVGNPVVTAIVFPYY370 pKa = 9.57FIIPIILLVAVGMGWYY386 pKa = 9.91DD387 pKa = 3.97FYY389 pKa = 11.89SEE391 pKa = 4.22TFYY394 pKa = 11.68SFVTWLVVPLLVLLTILAFVTATTLVVAIQANADD428 pKa = 3.36ICVSSGAPEE437 pKa = 4.16DD438 pKa = 4.35SILGVLNNLDD448 pKa = 4.56LEE450 pKa = 4.9DD451 pKa = 5.57DD452 pKa = 4.26SGSQSKK458 pKa = 10.81DD459 pKa = 3.28DD460 pKa = 4.91NGPSDD465 pKa = 3.52NFFYY469 pKa = 10.93DD470 pKa = 2.72IMVFYY475 pKa = 8.94THH477 pKa = 6.47QCTKK481 pKa = 9.45PNPWEE486 pKa = 4.0FMEE489 pKa = 4.5GHH491 pKa = 6.0YY492 pKa = 11.08ADD494 pKa = 4.74LARR497 pKa = 11.84GRR499 pKa = 11.84NILGEE504 pKa = 4.43FIRR507 pKa = 11.84SIEE510 pKa = 4.06DD511 pKa = 3.3TTLLEE516 pKa = 4.43LSQEE520 pKa = 4.48CGMEE524 pKa = 3.8YY525 pKa = 11.05GPIVVLLKK533 pKa = 10.68SLQDD537 pKa = 4.59LITILSQTSIRR548 pKa = 11.84ALTLMSCRR556 pKa = 11.84NVVPLYY562 pKa = 8.76TAWIYY567 pKa = 11.17EE568 pKa = 4.57STCTLAPVSLNLVFMCIVCIAFFGMVCITFRR599 pKa = 11.84GAYY602 pKa = 9.35YY603 pKa = 10.03PIDD606 pKa = 4.19FYY608 pKa = 11.76YY609 pKa = 11.06YY610 pKa = 10.35DD611 pKa = 3.3ATGRR615 pKa = 11.84GDD617 pKa = 3.52KK618 pKa = 10.69QEE620 pKa = 5.08LYY622 pKa = 8.74PTDD625 pKa = 3.58EE626 pKa = 4.52VGDD629 pKa = 3.97SGLADD634 pKa = 3.74TPCNEE639 pKa = 4.27KK640 pKa = 10.69VLEE643 pKa = 4.16EE644 pKa = 4.99QEE646 pKa = 4.36DD647 pKa = 3.75QCIDD651 pKa = 3.01ATEE654 pKa = 3.96AMEE657 pKa = 4.2EE658 pKa = 4.19SIEE661 pKa = 3.87IQIYY665 pKa = 10.33HH666 pKa = 7.27DD667 pKa = 4.51DD668 pKa = 3.8EE669 pKa = 4.5EE670 pKa = 5.75KK671 pKa = 10.66EE672 pKa = 4.15YY673 pKa = 9.86EE674 pKa = 4.18TDD676 pKa = 5.1DD677 pKa = 5.65DD678 pKa = 4.36EE679 pKa = 5.51QDD681 pKa = 3.3HH682 pKa = 6.55EE683 pKa = 5.72KK684 pKa = 11.21NFLLDD689 pKa = 3.65KK690 pKa = 10.67YY691 pKa = 9.36GEE693 pKa = 4.19PMEE696 pKa = 5.25GPDD699 pKa = 3.83AAQHH703 pKa = 5.65SVGTNGSYY711 pKa = 10.33RR712 pKa = 11.84GIDD715 pKa = 3.31RR716 pKa = 11.84NGDD719 pKa = 3.17SHH721 pKa = 7.78AVYY724 pKa = 10.74SDD726 pKa = 4.85DD727 pKa = 4.02DD728 pKa = 4.52DD729 pKa = 5.17EE730 pKa = 4.74MTFHH734 pKa = 6.58STFDD738 pKa = 3.91DD739 pKa = 4.17EE740 pKa = 5.16YY741 pKa = 9.5EE742 pKa = 4.24YY743 pKa = 11.69NSAAEE748 pKa = 4.32EE749 pKa = 3.97EE750 pKa = 4.95SEE752 pKa = 4.33TFSYY756 pKa = 11.28VSS758 pKa = 3.3
Molecular weight: 84.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A448ZSB5|A0A448ZSB5_9STRA Uncharacterized protein OS=Pseudo-nitzschia multistriata OX=183589 GN=PSNMU_V1.4_AUG-EV-PASAV3_0120360 PE=4 SV=1
MM1 pKa = 7.24VGAIPPGASGSLQNPLLPPHH21 pKa = 6.89RR22 pKa = 11.84SPWTMLATTGSPPRR36 pKa = 11.84ASGEE40 pKa = 3.95RR41 pKa = 11.84SSLVFDD47 pKa = 4.24RR48 pKa = 11.84RR49 pKa = 11.84ASILSEE55 pKa = 3.72GRR57 pKa = 11.84PPAFPVSLSMWPSRR71 pKa = 11.84CSSQKK76 pKa = 9.38SAQFSRR82 pKa = 11.84QGRR85 pKa = 11.84FGWKK89 pKa = 7.86VAPMVLSTGNPKK101 pKa = 10.18GASPFGDD108 pKa = 3.65TPALCIFASATPSSYY123 pKa = 10.64CPPGPLPQNSRR134 pKa = 11.84YY135 pKa = 9.89SKK137 pKa = 10.34RR138 pKa = 11.84AKK140 pKa = 9.71AWSLASCLPKK150 pKa = 10.61AIGLGTRR157 pKa = 11.84MVEE160 pKa = 4.14VSDD163 pKa = 3.71MALTPSYY170 pKa = 10.31SARR173 pKa = 11.84SMSYY177 pKa = 10.16PFRR180 pKa = 11.84ADD182 pKa = 2.91LRR184 pKa = 11.84KK185 pKa = 9.95YY186 pKa = 9.56RR187 pKa = 11.84VSVSASRR194 pKa = 11.84RR195 pKa = 11.84APVLAPPPPEE205 pKa = 4.36APPPVVGVSFTSLPRR220 pKa = 11.84VSDD223 pKa = 3.58AAVDD227 pKa = 3.85TVSRR231 pKa = 11.84KK232 pKa = 8.48VAAARR237 pKa = 11.84MAVEE241 pKa = 4.31GQPLVLLLMLLMLLLLLMLLPAAKK265 pKa = 10.14DD266 pKa = 3.31RR267 pKa = 11.84AGWAKK272 pKa = 10.57NPPPEE277 pKa = 4.24NADD280 pKa = 2.72ARR282 pKa = 11.84AAMEE286 pKa = 4.19APTRR290 pKa = 11.84SVLPDD295 pKa = 3.33RR296 pKa = 11.84LEE298 pKa = 4.07LKK300 pKa = 10.24PRR302 pKa = 11.84GRR304 pKa = 11.84TRR306 pKa = 11.84IMVCVRR312 pKa = 11.84FGLVLLAEE320 pKa = 4.72ASLEE324 pKa = 3.76RR325 pKa = 11.84SMYY328 pKa = 10.52SILRR332 pKa = 11.84CHH334 pKa = 6.62PNVPLRR340 pKa = 11.84CEE342 pKa = 3.77EE343 pKa = 4.07VLVVVTVPDD352 pKa = 3.59VTYY355 pKa = 10.63DD356 pKa = 2.77QYY358 pKa = 11.94DD359 pKa = 3.15RR360 pKa = 11.84HH361 pKa = 5.88RR362 pKa = 11.84RR363 pKa = 11.84NRR365 pKa = 11.84AKK367 pKa = 10.53SVFCCLEE374 pKa = 3.61QNPWIFSFKK383 pKa = 9.72MKK385 pKa = 9.61RR386 pKa = 11.84TRR388 pKa = 3.32
MM1 pKa = 7.24VGAIPPGASGSLQNPLLPPHH21 pKa = 6.89RR22 pKa = 11.84SPWTMLATTGSPPRR36 pKa = 11.84ASGEE40 pKa = 3.95RR41 pKa = 11.84SSLVFDD47 pKa = 4.24RR48 pKa = 11.84RR49 pKa = 11.84ASILSEE55 pKa = 3.72GRR57 pKa = 11.84PPAFPVSLSMWPSRR71 pKa = 11.84CSSQKK76 pKa = 9.38SAQFSRR82 pKa = 11.84QGRR85 pKa = 11.84FGWKK89 pKa = 7.86VAPMVLSTGNPKK101 pKa = 10.18GASPFGDD108 pKa = 3.65TPALCIFASATPSSYY123 pKa = 10.64CPPGPLPQNSRR134 pKa = 11.84YY135 pKa = 9.89SKK137 pKa = 10.34RR138 pKa = 11.84AKK140 pKa = 9.71AWSLASCLPKK150 pKa = 10.61AIGLGTRR157 pKa = 11.84MVEE160 pKa = 4.14VSDD163 pKa = 3.71MALTPSYY170 pKa = 10.31SARR173 pKa = 11.84SMSYY177 pKa = 10.16PFRR180 pKa = 11.84ADD182 pKa = 2.91LRR184 pKa = 11.84KK185 pKa = 9.95YY186 pKa = 9.56RR187 pKa = 11.84VSVSASRR194 pKa = 11.84RR195 pKa = 11.84APVLAPPPPEE205 pKa = 4.36APPPVVGVSFTSLPRR220 pKa = 11.84VSDD223 pKa = 3.58AAVDD227 pKa = 3.85TVSRR231 pKa = 11.84KK232 pKa = 8.48VAAARR237 pKa = 11.84MAVEE241 pKa = 4.31GQPLVLLLMLLMLLLLLMLLPAAKK265 pKa = 10.14DD266 pKa = 3.31RR267 pKa = 11.84AGWAKK272 pKa = 10.57NPPPEE277 pKa = 4.24NADD280 pKa = 2.72ARR282 pKa = 11.84AAMEE286 pKa = 4.19APTRR290 pKa = 11.84SVLPDD295 pKa = 3.33RR296 pKa = 11.84LEE298 pKa = 4.07LKK300 pKa = 10.24PRR302 pKa = 11.84GRR304 pKa = 11.84TRR306 pKa = 11.84IMVCVRR312 pKa = 11.84FGLVLLAEE320 pKa = 4.72ASLEE324 pKa = 3.76RR325 pKa = 11.84SMYY328 pKa = 10.52SILRR332 pKa = 11.84CHH334 pKa = 6.62PNVPLRR340 pKa = 11.84CEE342 pKa = 3.77EE343 pKa = 4.07VLVVVTVPDD352 pKa = 3.59VTYY355 pKa = 10.63DD356 pKa = 2.77QYY358 pKa = 11.94DD359 pKa = 3.15RR360 pKa = 11.84HH361 pKa = 5.88RR362 pKa = 11.84RR363 pKa = 11.84NRR365 pKa = 11.84AKK367 pKa = 10.53SVFCCLEE374 pKa = 3.61QNPWIFSFKK383 pKa = 9.72MKK385 pKa = 9.61RR386 pKa = 11.84TRR388 pKa = 3.32
Molecular weight: 42.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6325991 |
13 |
5812 |
531.3 |
58.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.042 ± 0.026 | 1.526 ± 0.009 |
6.31 ± 0.019 | 6.919 ± 0.025 |
3.46 ± 0.014 | 7.121 ± 0.026 |
2.241 ± 0.011 | 4.364 ± 0.014 |
5.531 ± 0.028 | 8.318 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.166 ± 0.009 | 4.428 ± 0.014 |
5.211 ± 0.021 | 3.68 ± 0.017 |
6.215 ± 0.022 | 8.918 ± 0.025 |
5.769 ± 0.014 | 5.95 ± 0.018 |
1.115 ± 0.007 | 2.393 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
