
Polaribacter sp. KT25b
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Polaribacter; unclassified Polaribacter
Average proteome isoelectric point is 6.89
Get precalculated fractions of proteins
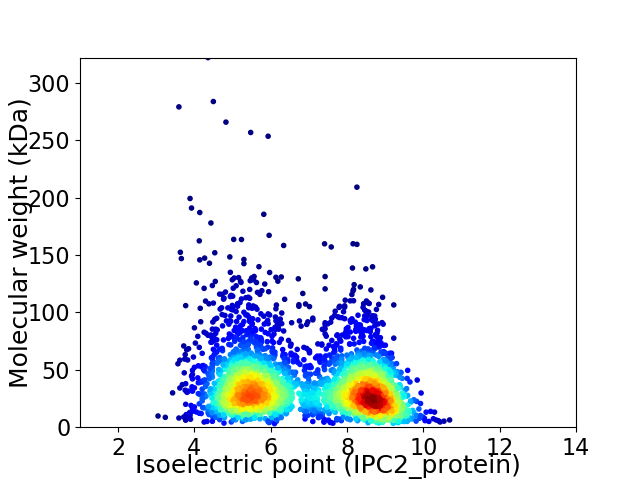
Virtual 2D-PAGE plot for 3350 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H1NZM2|A0A1H1NZM2_9FLAO Histidine phosphatase superfamily (Branch 1) OS=Polaribacter sp. KT25b OX=1855336 GN=SAMN05216503_1775 PE=4 SV=1
MM1 pKa = 7.36TKK3 pKa = 9.84IKK5 pKa = 10.65NILALIILATIVYY18 pKa = 9.87ACDD21 pKa = 3.51EE22 pKa = 4.33SSSVYY27 pKa = 10.45VDD29 pKa = 3.05NFDD32 pKa = 4.28YY33 pKa = 10.93EE34 pKa = 4.44GQALIDD40 pKa = 3.66SDD42 pKa = 4.28TLVKK46 pKa = 10.37FLSNHH51 pKa = 5.85YY52 pKa = 10.63YY53 pKa = 10.74DD54 pKa = 3.62VTEE57 pKa = 5.16DD58 pKa = 3.61EE59 pKa = 4.66VKK61 pKa = 10.94LLVSGKK67 pKa = 7.71TALIEE72 pKa = 4.57DD73 pKa = 4.55EE74 pKa = 5.12NLEE77 pKa = 4.15QYY79 pKa = 11.16DD80 pKa = 3.96VVDD83 pKa = 3.85NEE85 pKa = 3.71IDD87 pKa = 3.55YY88 pKa = 9.29TFYY91 pKa = 11.19YY92 pKa = 7.85YY93 pKa = 10.37TNRR96 pKa = 11.84VGSPTVEE103 pKa = 3.57KK104 pKa = 10.63GYY106 pKa = 8.3PTVMDD111 pKa = 3.93SVLVKK116 pKa = 10.73YY117 pKa = 9.86EE118 pKa = 3.76GKK120 pKa = 10.63RR121 pKa = 11.84IVNTDD126 pKa = 3.72SISASFDD133 pKa = 3.46KK134 pKa = 11.51NNGLWLTLNSVIRR147 pKa = 11.84GWTYY151 pKa = 10.2GIPKK155 pKa = 10.13FKK157 pKa = 10.9GGDD160 pKa = 3.52NITDD164 pKa = 3.77NGPITYY170 pKa = 9.97EE171 pKa = 3.88NGGKK175 pKa = 10.53GILFLPSGLAYY186 pKa = 10.16GPSGTTSILGSEE198 pKa = 4.14CLMFKK203 pKa = 10.29IEE205 pKa = 5.07LYY207 pKa = 11.04DD208 pKa = 4.26FIKK211 pKa = 10.54DD212 pKa = 3.46TDD214 pKa = 3.72HH215 pKa = 7.57DD216 pKa = 4.33NDD218 pKa = 3.77GVASFFEE225 pKa = 5.21DD226 pKa = 3.87PDD228 pKa = 5.02GDD230 pKa = 3.9GDD232 pKa = 4.44PRR234 pKa = 11.84NDD236 pKa = 3.71DD237 pKa = 3.52TDD239 pKa = 5.26LDD241 pKa = 4.14GSPNYY246 pKa = 10.8ADD248 pKa = 5.37ADD250 pKa = 4.09DD251 pKa = 5.71DD252 pKa = 4.92GDD254 pKa = 4.86GILTINEE261 pKa = 4.24DD262 pKa = 4.01ANSDD266 pKa = 3.77GNPANDD272 pKa = 4.89FSDD275 pKa = 4.18PDD277 pKa = 3.61NSTLADD283 pKa = 3.84YY284 pKa = 11.07LNSLIKK290 pKa = 10.73VSNN293 pKa = 3.62
MM1 pKa = 7.36TKK3 pKa = 9.84IKK5 pKa = 10.65NILALIILATIVYY18 pKa = 9.87ACDD21 pKa = 3.51EE22 pKa = 4.33SSSVYY27 pKa = 10.45VDD29 pKa = 3.05NFDD32 pKa = 4.28YY33 pKa = 10.93EE34 pKa = 4.44GQALIDD40 pKa = 3.66SDD42 pKa = 4.28TLVKK46 pKa = 10.37FLSNHH51 pKa = 5.85YY52 pKa = 10.63YY53 pKa = 10.74DD54 pKa = 3.62VTEE57 pKa = 5.16DD58 pKa = 3.61EE59 pKa = 4.66VKK61 pKa = 10.94LLVSGKK67 pKa = 7.71TALIEE72 pKa = 4.57DD73 pKa = 4.55EE74 pKa = 5.12NLEE77 pKa = 4.15QYY79 pKa = 11.16DD80 pKa = 3.96VVDD83 pKa = 3.85NEE85 pKa = 3.71IDD87 pKa = 3.55YY88 pKa = 9.29TFYY91 pKa = 11.19YY92 pKa = 7.85YY93 pKa = 10.37TNRR96 pKa = 11.84VGSPTVEE103 pKa = 3.57KK104 pKa = 10.63GYY106 pKa = 8.3PTVMDD111 pKa = 3.93SVLVKK116 pKa = 10.73YY117 pKa = 9.86EE118 pKa = 3.76GKK120 pKa = 10.63RR121 pKa = 11.84IVNTDD126 pKa = 3.72SISASFDD133 pKa = 3.46KK134 pKa = 11.51NNGLWLTLNSVIRR147 pKa = 11.84GWTYY151 pKa = 10.2GIPKK155 pKa = 10.13FKK157 pKa = 10.9GGDD160 pKa = 3.52NITDD164 pKa = 3.77NGPITYY170 pKa = 9.97EE171 pKa = 3.88NGGKK175 pKa = 10.53GILFLPSGLAYY186 pKa = 10.16GPSGTTSILGSEE198 pKa = 4.14CLMFKK203 pKa = 10.29IEE205 pKa = 5.07LYY207 pKa = 11.04DD208 pKa = 4.26FIKK211 pKa = 10.54DD212 pKa = 3.46TDD214 pKa = 3.72HH215 pKa = 7.57DD216 pKa = 4.33NDD218 pKa = 3.77GVASFFEE225 pKa = 5.21DD226 pKa = 3.87PDD228 pKa = 5.02GDD230 pKa = 3.9GDD232 pKa = 4.44PRR234 pKa = 11.84NDD236 pKa = 3.71DD237 pKa = 3.52TDD239 pKa = 5.26LDD241 pKa = 4.14GSPNYY246 pKa = 10.8ADD248 pKa = 5.37ADD250 pKa = 4.09DD251 pKa = 5.71DD252 pKa = 4.92GDD254 pKa = 4.86GILTINEE261 pKa = 4.24DD262 pKa = 4.01ANSDD266 pKa = 3.77GNPANDD272 pKa = 4.89FSDD275 pKa = 4.18PDD277 pKa = 3.61NSTLADD283 pKa = 3.84YY284 pKa = 11.07LNSLIKK290 pKa = 10.73VSNN293 pKa = 3.62
Molecular weight: 32.22 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H1RSI1|A0A1H1RSI1_9FLAO Uncharacterized protein OS=Polaribacter sp. KT25b OX=1855336 GN=SAMN05216503_2899 PE=4 SV=1
MM1 pKa = 7.66ASLIGHH7 pKa = 7.75AFTSIALGKK16 pKa = 10.04SFSRR20 pKa = 11.84NRR22 pKa = 11.84RR23 pKa = 11.84NWKK26 pKa = 9.89LILIAIVCAIIQDD39 pKa = 3.1IDD41 pKa = 3.9VIGFQFGIPYY51 pKa = 10.22NSFWGHH57 pKa = 6.29RR58 pKa = 11.84GFTHH62 pKa = 6.91SLFFALLLAILVTAIFYY79 pKa = 10.41KK80 pKa = 10.8KK81 pKa = 10.61LFFSKK86 pKa = 10.34KK87 pKa = 10.2GIILILFFFLCGAFHH102 pKa = 7.39SILDD106 pKa = 3.75AFTDD110 pKa = 3.85GGNGVAFFTPFNNTRR125 pKa = 11.84YY126 pKa = 8.83FFPWRR131 pKa = 11.84PIKK134 pKa = 10.48ASPLGITRR142 pKa = 11.84FFSEE146 pKa = 3.98RR147 pKa = 11.84GMLVIRR153 pKa = 11.84SEE155 pKa = 4.4VTWIVLPGFIYY166 pKa = 9.62MVIAFLIRR174 pKa = 11.84KK175 pKa = 8.54KK176 pKa = 10.51RR177 pKa = 11.84NKK179 pKa = 9.51II180 pKa = 3.38
MM1 pKa = 7.66ASLIGHH7 pKa = 7.75AFTSIALGKK16 pKa = 10.04SFSRR20 pKa = 11.84NRR22 pKa = 11.84RR23 pKa = 11.84NWKK26 pKa = 9.89LILIAIVCAIIQDD39 pKa = 3.1IDD41 pKa = 3.9VIGFQFGIPYY51 pKa = 10.22NSFWGHH57 pKa = 6.29RR58 pKa = 11.84GFTHH62 pKa = 6.91SLFFALLLAILVTAIFYY79 pKa = 10.41KK80 pKa = 10.8KK81 pKa = 10.61LFFSKK86 pKa = 10.34KK87 pKa = 10.2GIILILFFFLCGAFHH102 pKa = 7.39SILDD106 pKa = 3.75AFTDD110 pKa = 3.85GGNGVAFFTPFNNTRR125 pKa = 11.84YY126 pKa = 8.83FFPWRR131 pKa = 11.84PIKK134 pKa = 10.48ASPLGITRR142 pKa = 11.84FFSEE146 pKa = 3.98RR147 pKa = 11.84GMLVIRR153 pKa = 11.84SEE155 pKa = 4.4VTWIVLPGFIYY166 pKa = 9.62MVIAFLIRR174 pKa = 11.84KK175 pKa = 8.54KK176 pKa = 10.51RR177 pKa = 11.84NKK179 pKa = 9.51II180 pKa = 3.38
Molecular weight: 20.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1171335 |
29 |
3002 |
349.7 |
39.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.899 ± 0.036 | 0.678 ± 0.012 |
5.502 ± 0.035 | 6.602 ± 0.037 |
5.532 ± 0.039 | 5.995 ± 0.039 |
1.602 ± 0.017 | 8.566 ± 0.043 |
8.864 ± 0.055 | 9.167 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.984 ± 0.022 | 6.824 ± 0.045 |
3.068 ± 0.023 | 3.09 ± 0.02 |
2.997 ± 0.022 | 6.68 ± 0.039 |
5.946 ± 0.042 | 5.949 ± 0.031 |
1.031 ± 0.015 | 4.023 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
