
Giraffa camelopardalis papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Deltapapillomavirus; Deltapapillomavirus 7
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3GV90|A0A1L3GV90_9PAPI E5 protein OS=Giraffa camelopardalis papillomavirus 1 OX=1922325 GN=E5 PE=4 SV=1
MM1 pKa = 7.51ASVRR5 pKa = 11.84RR6 pKa = 11.84VKK8 pKa = 10.41RR9 pKa = 11.84ASAADD14 pKa = 4.16LYY16 pKa = 9.65RR17 pKa = 11.84TCKK20 pKa = 10.51ASNTCPPDD28 pKa = 3.74VIPKK32 pKa = 9.92IEE34 pKa = 4.35GSTIADD40 pKa = 4.11KK41 pKa = 10.74ILQYY45 pKa = 11.28GSLGVFFGGLGIGTGRR61 pKa = 11.84GSAAVVPGVTVPKK74 pKa = 10.13LVSRR78 pKa = 11.84GGGYY82 pKa = 9.79QPLNTSSSSISLTSTGSVRR101 pKa = 11.84GVAARR106 pKa = 11.84LRR108 pKa = 11.84EE109 pKa = 4.23FNVLPPATEE118 pKa = 4.21VIPLEE123 pKa = 4.33TFASGGAEE131 pKa = 4.15GISPSVEE138 pKa = 3.85LVPEE142 pKa = 4.34SSSILTPEE150 pKa = 3.98NGLAEE155 pKa = 4.28TALGAEE161 pKa = 4.55VFADD165 pKa = 3.56TDD167 pKa = 3.62TTLITLHH174 pKa = 6.91SDD176 pKa = 3.63TGPGDD181 pKa = 3.62VAVLEE186 pKa = 4.35LRR188 pKa = 11.84PTEE191 pKa = 4.36NDD193 pKa = 3.33LQAHH197 pKa = 5.79TLTRR201 pKa = 11.84GPSAHH206 pKa = 6.23TPSFVDD212 pKa = 3.07THH214 pKa = 8.19AVMFGEE220 pKa = 4.68TSSSEE225 pKa = 3.78NVFIGGANVGNVEE238 pKa = 4.16GEE240 pKa = 4.26FIEE243 pKa = 5.72LSNLDD248 pKa = 4.08PRR250 pKa = 11.84TSTPEE255 pKa = 3.83GEE257 pKa = 4.47ARR259 pKa = 11.84PPARR263 pKa = 11.84GRR265 pKa = 11.84GVWNWFSKK273 pKa = 10.22RR274 pKa = 11.84YY275 pKa = 8.81YY276 pKa = 9.92SQVPVTDD283 pKa = 3.95PDD285 pKa = 4.07FVSSYY290 pKa = 9.39TFEE293 pKa = 4.35NPLYY297 pKa = 9.44EE298 pKa = 4.35YY299 pKa = 11.19ADD301 pKa = 3.91GVQTGQSFPTVSNQGRR317 pKa = 11.84VGISRR322 pKa = 11.84LGWQEE327 pKa = 3.81GVSTRR332 pKa = 11.84SGAQVGRR339 pKa = 11.84ALHH342 pKa = 6.0LRR344 pKa = 11.84YY345 pKa = 9.9SLSSIEE351 pKa = 4.27NPPVDD356 pKa = 4.9VIPLSISAGEE366 pKa = 4.15EE367 pKa = 3.86GTLQQEE373 pKa = 4.41VSFQVSDD380 pKa = 3.7DD381 pKa = 3.8TFDD384 pKa = 3.65TVDD387 pKa = 3.53LFEE390 pKa = 7.32DD391 pKa = 3.64EE392 pKa = 5.02DD393 pKa = 4.89APLLRR398 pKa = 11.84TEE400 pKa = 3.93QHH402 pKa = 5.13TLRR405 pKa = 11.84TGSRR409 pKa = 11.84RR410 pKa = 11.84NTVTPVLHH418 pKa = 6.73RR419 pKa = 11.84PHH421 pKa = 6.86IAGATVFDD429 pKa = 4.27KK430 pKa = 11.32AVGVKK435 pKa = 10.17GQGGSSTVPVDD446 pKa = 3.13TDD448 pKa = 3.25GGVIVDD454 pKa = 4.11HH455 pKa = 7.65DD456 pKa = 4.89DD457 pKa = 3.69YY458 pKa = 11.91VPTSTPTIIIDD469 pKa = 3.44NEE471 pKa = 3.98LAVWFHH477 pKa = 7.32SYY479 pKa = 10.37FLHH482 pKa = 6.86PSKK485 pKa = 10.59LGKK488 pKa = 9.86RR489 pKa = 11.84KK490 pKa = 9.84RR491 pKa = 11.84KK492 pKa = 10.0RR493 pKa = 11.84SDD495 pKa = 2.98SSVV498 pKa = 2.79
MM1 pKa = 7.51ASVRR5 pKa = 11.84RR6 pKa = 11.84VKK8 pKa = 10.41RR9 pKa = 11.84ASAADD14 pKa = 4.16LYY16 pKa = 9.65RR17 pKa = 11.84TCKK20 pKa = 10.51ASNTCPPDD28 pKa = 3.74VIPKK32 pKa = 9.92IEE34 pKa = 4.35GSTIADD40 pKa = 4.11KK41 pKa = 10.74ILQYY45 pKa = 11.28GSLGVFFGGLGIGTGRR61 pKa = 11.84GSAAVVPGVTVPKK74 pKa = 10.13LVSRR78 pKa = 11.84GGGYY82 pKa = 9.79QPLNTSSSSISLTSTGSVRR101 pKa = 11.84GVAARR106 pKa = 11.84LRR108 pKa = 11.84EE109 pKa = 4.23FNVLPPATEE118 pKa = 4.21VIPLEE123 pKa = 4.33TFASGGAEE131 pKa = 4.15GISPSVEE138 pKa = 3.85LVPEE142 pKa = 4.34SSSILTPEE150 pKa = 3.98NGLAEE155 pKa = 4.28TALGAEE161 pKa = 4.55VFADD165 pKa = 3.56TDD167 pKa = 3.62TTLITLHH174 pKa = 6.91SDD176 pKa = 3.63TGPGDD181 pKa = 3.62VAVLEE186 pKa = 4.35LRR188 pKa = 11.84PTEE191 pKa = 4.36NDD193 pKa = 3.33LQAHH197 pKa = 5.79TLTRR201 pKa = 11.84GPSAHH206 pKa = 6.23TPSFVDD212 pKa = 3.07THH214 pKa = 8.19AVMFGEE220 pKa = 4.68TSSSEE225 pKa = 3.78NVFIGGANVGNVEE238 pKa = 4.16GEE240 pKa = 4.26FIEE243 pKa = 5.72LSNLDD248 pKa = 4.08PRR250 pKa = 11.84TSTPEE255 pKa = 3.83GEE257 pKa = 4.47ARR259 pKa = 11.84PPARR263 pKa = 11.84GRR265 pKa = 11.84GVWNWFSKK273 pKa = 10.22RR274 pKa = 11.84YY275 pKa = 8.81YY276 pKa = 9.92SQVPVTDD283 pKa = 3.95PDD285 pKa = 4.07FVSSYY290 pKa = 9.39TFEE293 pKa = 4.35NPLYY297 pKa = 9.44EE298 pKa = 4.35YY299 pKa = 11.19ADD301 pKa = 3.91GVQTGQSFPTVSNQGRR317 pKa = 11.84VGISRR322 pKa = 11.84LGWQEE327 pKa = 3.81GVSTRR332 pKa = 11.84SGAQVGRR339 pKa = 11.84ALHH342 pKa = 6.0LRR344 pKa = 11.84YY345 pKa = 9.9SLSSIEE351 pKa = 4.27NPPVDD356 pKa = 4.9VIPLSISAGEE366 pKa = 4.15EE367 pKa = 3.86GTLQQEE373 pKa = 4.41VSFQVSDD380 pKa = 3.7DD381 pKa = 3.8TFDD384 pKa = 3.65TVDD387 pKa = 3.53LFEE390 pKa = 7.32DD391 pKa = 3.64EE392 pKa = 5.02DD393 pKa = 4.89APLLRR398 pKa = 11.84TEE400 pKa = 3.93QHH402 pKa = 5.13TLRR405 pKa = 11.84TGSRR409 pKa = 11.84RR410 pKa = 11.84NTVTPVLHH418 pKa = 6.73RR419 pKa = 11.84PHH421 pKa = 6.86IAGATVFDD429 pKa = 4.27KK430 pKa = 11.32AVGVKK435 pKa = 10.17GQGGSSTVPVDD446 pKa = 3.13TDD448 pKa = 3.25GGVIVDD454 pKa = 4.11HH455 pKa = 7.65DD456 pKa = 4.89DD457 pKa = 3.69YY458 pKa = 11.91VPTSTPTIIIDD469 pKa = 3.44NEE471 pKa = 3.98LAVWFHH477 pKa = 7.32SYY479 pKa = 10.37FLHH482 pKa = 6.86PSKK485 pKa = 10.59LGKK488 pKa = 9.86RR489 pKa = 11.84KK490 pKa = 9.84RR491 pKa = 11.84KK492 pKa = 10.0RR493 pKa = 11.84SDD495 pKa = 2.98SSVV498 pKa = 2.79
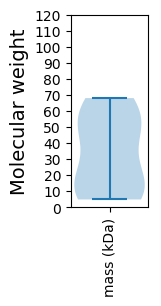
Molecular weight: 52.98 kDa
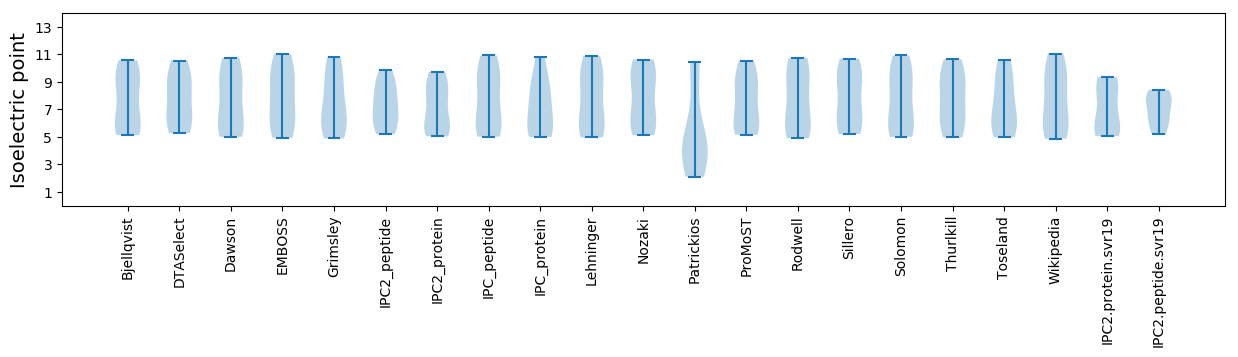
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3GV75|A0A1L3GV75_9PAPI Regulatory protein E2 OS=Giraffa camelopardalis papillomavirus 1 OX=1922325 GN=E2 PE=3 SV=1
DDD2 pKa = 4.31KKK4 pKa = 11.09DD5 pKa = 3.42LIILILRR12 pKa = 11.84LPEEE16 pKa = 4.07NLTDDD21 pKa = 3.53TSRR24 pKa = 11.84TVTQEEE30 pKa = 3.72DD31 pKa = 4.24QPTQPSLSLLEEE43 pKa = 4.3KKK45 pKa = 9.46QQVALSEEE53 pKa = 4.16EE54 pKa = 4.19AVGYYY59 pKa = 7.28VCLARR64 pKa = 11.84TPTIYYY70 pKa = 10.05GPRR73 pKa = 11.84GTHHH77 pKa = 6.03HH78 pKa = 7.31YYY80 pKa = 10.97PPPRR84 pKa = 11.84CRR86 pKa = 11.84ARR88 pKa = 11.84YYY90 pKa = 8.21WTWHHH95 pKa = 4.25VRR97 pKa = 11.84KKK99 pKa = 9.48HH100 pKa = 5.06RR101 pKa = 11.84PRR103 pKa = 11.84RR104 pKa = 11.84ARR106 pKa = 11.84IPRR109 pKa = 11.84S
DDD2 pKa = 4.31KKK4 pKa = 11.09DD5 pKa = 3.42LIILILRR12 pKa = 11.84LPEEE16 pKa = 4.07NLTDDD21 pKa = 3.53TSRR24 pKa = 11.84TVTQEEE30 pKa = 3.72DD31 pKa = 4.24QPTQPSLSLLEEE43 pKa = 4.3KKK45 pKa = 9.46QQVALSEEE53 pKa = 4.16EE54 pKa = 4.19AVGYYY59 pKa = 7.28VCLARR64 pKa = 11.84TPTIYYY70 pKa = 10.05GPRR73 pKa = 11.84GTHHH77 pKa = 6.03HH78 pKa = 7.31YYY80 pKa = 10.97PPPRR84 pKa = 11.84CRR86 pKa = 11.84ARR88 pKa = 11.84YYY90 pKa = 8.21WTWHHH95 pKa = 4.25VRR97 pKa = 11.84KKK99 pKa = 9.48HH100 pKa = 5.06RR101 pKa = 11.84PRR103 pKa = 11.84RR104 pKa = 11.84ARR106 pKa = 11.84IPRR109 pKa = 11.84S
Molecular weight: 12.78 kDa
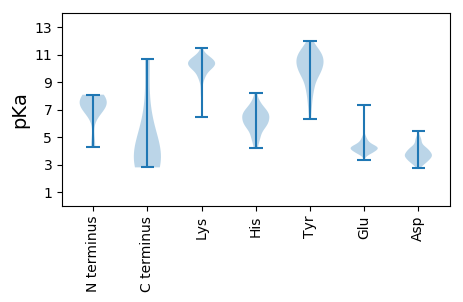
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2411 |
41 |
609 |
301.4 |
33.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.341 ± 0.67 | 2.696 ± 0.632 |
5.185 ± 0.404 | 5.599 ± 0.322 |
3.857 ± 0.595 | 7.217 ± 0.768 |
2.655 ± 0.236 | 3.401 ± 0.455 |
5.019 ± 0.757 | 8.752 ± 0.672 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.701 ± 0.405 | 3.608 ± 0.627 |
6.429 ± 0.637 | 4.521 ± 0.633 |
5.973 ± 0.782 | 7.756 ± 0.682 |
6.678 ± 0.667 | 7.01 ± 0.618 |
1.493 ± 0.24 | 3.111 ± 0.395 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
