
Capybara microvirus Cap1_SP_77
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
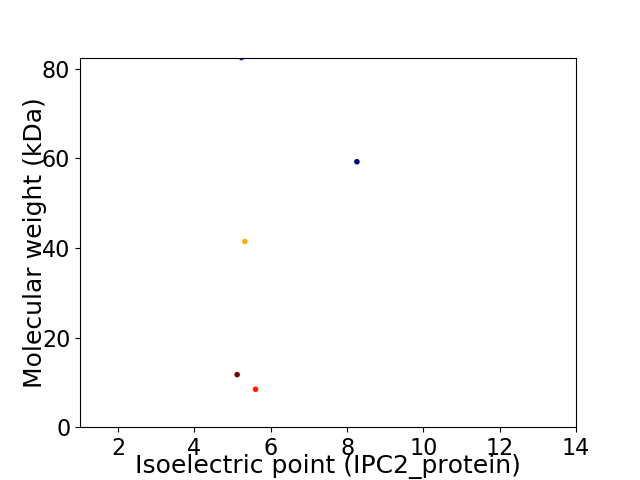
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>tr|A0A4V1FVN1|A0A4V1FVN1_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_77 OX=2584791 PE=4 SV=1
MM1 pKa = 7.46SNVFIEE7 pKa = 4.7RR8 pKa = 11.84EE9 pKa = 4.01NFSPNIKK16 pKa = 10.31RR17 pKa = 11.84NVFDD21 pKa = 5.95LSHH24 pKa = 6.29QNNLSLKK31 pKa = 9.12IGKK34 pKa = 8.0ITPIYY39 pKa = 8.86CQEE42 pKa = 4.08VLPGTSVKK50 pKa = 9.85MDD52 pKa = 3.24VVSALRR58 pKa = 11.84AAPLFFPIQTKK69 pKa = 10.17IKK71 pKa = 9.69ASMSFFYY78 pKa = 10.89VRR80 pKa = 11.84NRR82 pKa = 11.84NLWDD86 pKa = 3.62DD87 pKa = 3.31WTKK90 pKa = 10.78FISNEE95 pKa = 4.17EE96 pKa = 4.12TSSTFPVIDD105 pKa = 3.62ASKK108 pKa = 10.12NPEE111 pKa = 3.83FFKK114 pKa = 11.24VGGLADD120 pKa = 3.86YY121 pKa = 10.93LGVPTAFYY129 pKa = 10.16GYY131 pKa = 7.09PTASLTTALDD141 pKa = 3.65AEE143 pKa = 4.6EE144 pKa = 4.16VLSNLEE150 pKa = 3.5TSYY153 pKa = 11.51YY154 pKa = 9.46EE155 pKa = 4.29KK156 pKa = 10.73KK157 pKa = 10.62KK158 pKa = 9.27STHH161 pKa = 5.21SLAYY165 pKa = 8.99VQHH168 pKa = 6.71SYY170 pKa = 11.25DD171 pKa = 3.44VSSDD175 pKa = 3.24RR176 pKa = 11.84MAGYY180 pKa = 10.3CGIINSTITPGVKK193 pKa = 9.99FSAVSFGGNRR203 pKa = 11.84SGEE206 pKa = 4.44SVVGNYY212 pKa = 9.16DD213 pKa = 3.85LPWTAGGFTYY223 pKa = 10.79NKK225 pKa = 8.29YY226 pKa = 9.48GRR228 pKa = 11.84VYY230 pKa = 10.67RR231 pKa = 11.84VTKK234 pKa = 10.45KK235 pKa = 10.32LASGNMTVKK244 pKa = 10.38SRR246 pKa = 11.84SACSLMVIVNGTVVAVQTGSYY267 pKa = 10.37ASSTTTIGLLQTVVDD282 pKa = 5.84AINKK286 pKa = 10.03AIDD289 pKa = 4.09DD290 pKa = 4.15GQVVHH295 pKa = 6.75FCVFTQGTQTSFSPSTIYY313 pKa = 10.79YY314 pKa = 9.29IEE316 pKa = 3.98QLIVQYY322 pKa = 11.31DD323 pKa = 3.7NTDD326 pKa = 3.11ATEE329 pKa = 3.82WKK331 pKa = 10.23AISGYY336 pKa = 10.56NSYY339 pKa = 11.45NPFLADD345 pKa = 3.61GMKK348 pKa = 10.57LSALPFRR355 pKa = 11.84AYY357 pKa = 10.18EE358 pKa = 3.99SVYY361 pKa = 9.5NCFYY365 pKa = 10.45RR366 pKa = 11.84KK367 pKa = 9.51EE368 pKa = 4.12RR369 pKa = 11.84VDD371 pKa = 3.54PNRR374 pKa = 11.84VNGKK378 pKa = 9.11IEE380 pKa = 3.97YY381 pKa = 9.94DD382 pKa = 3.91VYY384 pKa = 11.12CDD386 pKa = 3.76KK387 pKa = 11.58SGGNDD392 pKa = 2.82TNYY395 pKa = 10.22YY396 pKa = 9.93HH397 pKa = 7.48LYY399 pKa = 9.2DD400 pKa = 4.93APWEE404 pKa = 4.15TDD406 pKa = 2.84QFTSCLPTPQAGVAPLVGLTSNFSGVVSATFLDD439 pKa = 4.12EE440 pKa = 4.33SGNTVKK446 pKa = 10.85YY447 pKa = 10.32DD448 pKa = 3.59VQSVTDD454 pKa = 3.7SSGKK458 pKa = 8.01ITGFAYY464 pKa = 10.58SEE466 pKa = 4.85DD467 pKa = 3.95IPWSVRR473 pKa = 11.84EE474 pKa = 3.96QLNHH478 pKa = 6.04SVLSGFDD485 pKa = 3.28INSLRR490 pKa = 11.84NVSAFQIYY498 pKa = 10.45LEE500 pKa = 4.29NKK502 pKa = 8.66LRR504 pKa = 11.84RR505 pKa = 11.84GLKK508 pKa = 10.09YY509 pKa = 9.42VDD511 pKa = 3.64QIKK514 pKa = 10.15SHH516 pKa = 6.78FGVQPSYY523 pKa = 10.89EE524 pKa = 4.09ALNMPEE530 pKa = 4.11YY531 pKa = 10.32IGGVTEE537 pKa = 4.27TFNVNQISSTIEE549 pKa = 3.57NDD551 pKa = 3.33KK552 pKa = 11.23VSLGDD557 pKa = 3.33YY558 pKa = 9.63AGQMTCMAHH567 pKa = 6.24PNHH570 pKa = 6.67NISHH574 pKa = 6.14YY575 pKa = 9.83CDD577 pKa = 2.98EE578 pKa = 4.89HH579 pKa = 7.76GFIIGVLSIVPIPNYY594 pKa = 10.41SQALPKK600 pKa = 10.59FFGKK604 pKa = 7.32TTTFDD609 pKa = 3.83YY610 pKa = 8.18YY611 pKa = 10.93TPEE614 pKa = 4.25LDD616 pKa = 4.3KK617 pKa = 11.56LSLQPVRR624 pKa = 11.84NWEE627 pKa = 3.88IAPVQTFANADD638 pKa = 3.52SGSKK642 pKa = 9.72IEE644 pKa = 4.6ALDD647 pKa = 3.76GVFGYY652 pKa = 9.28QLPWYY657 pKa = 8.36EE658 pKa = 4.01YY659 pKa = 10.5KK660 pKa = 10.9GALDD664 pKa = 3.81TVHH667 pKa = 8.04GEE669 pKa = 3.68MRR671 pKa = 11.84TDD673 pKa = 3.76LRR675 pKa = 11.84DD676 pKa = 3.52FMMNRR681 pKa = 11.84VFRR684 pKa = 11.84TTPQLGSDD692 pKa = 3.8FVYY695 pKa = 11.14VDD697 pKa = 3.75GEE699 pKa = 4.55SVNNVFNVQDD709 pKa = 4.14DD710 pKa = 4.51DD711 pKa = 3.89VDD713 pKa = 4.14KK714 pKa = 10.88IYY716 pKa = 10.73GQIYY720 pKa = 9.59FDD722 pKa = 4.28VKK724 pKa = 9.6TKK726 pKa = 10.88HH727 pKa = 5.21PMSRR731 pKa = 11.84IGQPVYY737 pKa = 10.36HH738 pKa = 7.06
MM1 pKa = 7.46SNVFIEE7 pKa = 4.7RR8 pKa = 11.84EE9 pKa = 4.01NFSPNIKK16 pKa = 10.31RR17 pKa = 11.84NVFDD21 pKa = 5.95LSHH24 pKa = 6.29QNNLSLKK31 pKa = 9.12IGKK34 pKa = 8.0ITPIYY39 pKa = 8.86CQEE42 pKa = 4.08VLPGTSVKK50 pKa = 9.85MDD52 pKa = 3.24VVSALRR58 pKa = 11.84AAPLFFPIQTKK69 pKa = 10.17IKK71 pKa = 9.69ASMSFFYY78 pKa = 10.89VRR80 pKa = 11.84NRR82 pKa = 11.84NLWDD86 pKa = 3.62DD87 pKa = 3.31WTKK90 pKa = 10.78FISNEE95 pKa = 4.17EE96 pKa = 4.12TSSTFPVIDD105 pKa = 3.62ASKK108 pKa = 10.12NPEE111 pKa = 3.83FFKK114 pKa = 11.24VGGLADD120 pKa = 3.86YY121 pKa = 10.93LGVPTAFYY129 pKa = 10.16GYY131 pKa = 7.09PTASLTTALDD141 pKa = 3.65AEE143 pKa = 4.6EE144 pKa = 4.16VLSNLEE150 pKa = 3.5TSYY153 pKa = 11.51YY154 pKa = 9.46EE155 pKa = 4.29KK156 pKa = 10.73KK157 pKa = 10.62KK158 pKa = 9.27STHH161 pKa = 5.21SLAYY165 pKa = 8.99VQHH168 pKa = 6.71SYY170 pKa = 11.25DD171 pKa = 3.44VSSDD175 pKa = 3.24RR176 pKa = 11.84MAGYY180 pKa = 10.3CGIINSTITPGVKK193 pKa = 9.99FSAVSFGGNRR203 pKa = 11.84SGEE206 pKa = 4.44SVVGNYY212 pKa = 9.16DD213 pKa = 3.85LPWTAGGFTYY223 pKa = 10.79NKK225 pKa = 8.29YY226 pKa = 9.48GRR228 pKa = 11.84VYY230 pKa = 10.67RR231 pKa = 11.84VTKK234 pKa = 10.45KK235 pKa = 10.32LASGNMTVKK244 pKa = 10.38SRR246 pKa = 11.84SACSLMVIVNGTVVAVQTGSYY267 pKa = 10.37ASSTTTIGLLQTVVDD282 pKa = 5.84AINKK286 pKa = 10.03AIDD289 pKa = 4.09DD290 pKa = 4.15GQVVHH295 pKa = 6.75FCVFTQGTQTSFSPSTIYY313 pKa = 10.79YY314 pKa = 9.29IEE316 pKa = 3.98QLIVQYY322 pKa = 11.31DD323 pKa = 3.7NTDD326 pKa = 3.11ATEE329 pKa = 3.82WKK331 pKa = 10.23AISGYY336 pKa = 10.56NSYY339 pKa = 11.45NPFLADD345 pKa = 3.61GMKK348 pKa = 10.57LSALPFRR355 pKa = 11.84AYY357 pKa = 10.18EE358 pKa = 3.99SVYY361 pKa = 9.5NCFYY365 pKa = 10.45RR366 pKa = 11.84KK367 pKa = 9.51EE368 pKa = 4.12RR369 pKa = 11.84VDD371 pKa = 3.54PNRR374 pKa = 11.84VNGKK378 pKa = 9.11IEE380 pKa = 3.97YY381 pKa = 9.94DD382 pKa = 3.91VYY384 pKa = 11.12CDD386 pKa = 3.76KK387 pKa = 11.58SGGNDD392 pKa = 2.82TNYY395 pKa = 10.22YY396 pKa = 9.93HH397 pKa = 7.48LYY399 pKa = 9.2DD400 pKa = 4.93APWEE404 pKa = 4.15TDD406 pKa = 2.84QFTSCLPTPQAGVAPLVGLTSNFSGVVSATFLDD439 pKa = 4.12EE440 pKa = 4.33SGNTVKK446 pKa = 10.85YY447 pKa = 10.32DD448 pKa = 3.59VQSVTDD454 pKa = 3.7SSGKK458 pKa = 8.01ITGFAYY464 pKa = 10.58SEE466 pKa = 4.85DD467 pKa = 3.95IPWSVRR473 pKa = 11.84EE474 pKa = 3.96QLNHH478 pKa = 6.04SVLSGFDD485 pKa = 3.28INSLRR490 pKa = 11.84NVSAFQIYY498 pKa = 10.45LEE500 pKa = 4.29NKK502 pKa = 8.66LRR504 pKa = 11.84RR505 pKa = 11.84GLKK508 pKa = 10.09YY509 pKa = 9.42VDD511 pKa = 3.64QIKK514 pKa = 10.15SHH516 pKa = 6.78FGVQPSYY523 pKa = 10.89EE524 pKa = 4.09ALNMPEE530 pKa = 4.11YY531 pKa = 10.32IGGVTEE537 pKa = 4.27TFNVNQISSTIEE549 pKa = 3.57NDD551 pKa = 3.33KK552 pKa = 11.23VSLGDD557 pKa = 3.33YY558 pKa = 9.63AGQMTCMAHH567 pKa = 6.24PNHH570 pKa = 6.67NISHH574 pKa = 6.14YY575 pKa = 9.83CDD577 pKa = 2.98EE578 pKa = 4.89HH579 pKa = 7.76GFIIGVLSIVPIPNYY594 pKa = 10.41SQALPKK600 pKa = 10.59FFGKK604 pKa = 7.32TTTFDD609 pKa = 3.83YY610 pKa = 8.18YY611 pKa = 10.93TPEE614 pKa = 4.25LDD616 pKa = 4.3KK617 pKa = 11.56LSLQPVRR624 pKa = 11.84NWEE627 pKa = 3.88IAPVQTFANADD638 pKa = 3.52SGSKK642 pKa = 9.72IEE644 pKa = 4.6ALDD647 pKa = 3.76GVFGYY652 pKa = 9.28QLPWYY657 pKa = 8.36EE658 pKa = 4.01YY659 pKa = 10.5KK660 pKa = 10.9GALDD664 pKa = 3.81TVHH667 pKa = 8.04GEE669 pKa = 3.68MRR671 pKa = 11.84TDD673 pKa = 3.76LRR675 pKa = 11.84DD676 pKa = 3.52FMMNRR681 pKa = 11.84VFRR684 pKa = 11.84TTPQLGSDD692 pKa = 3.8FVYY695 pKa = 11.14VDD697 pKa = 3.75GEE699 pKa = 4.55SVNNVFNVQDD709 pKa = 4.14DD710 pKa = 4.51DD711 pKa = 3.89VDD713 pKa = 4.14KK714 pKa = 10.88IYY716 pKa = 10.73GQIYY720 pKa = 9.59FDD722 pKa = 4.28VKK724 pKa = 9.6TKK726 pKa = 10.88HH727 pKa = 5.21PMSRR731 pKa = 11.84IGQPVYY737 pKa = 10.36HH738 pKa = 7.06
Molecular weight: 82.47 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4R6|A0A4P8W4R6_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_77 OX=2584791 PE=3 SV=1
MM1 pKa = 7.51KK2 pKa = 10.17CLNPVIMINRR12 pKa = 11.84PFLYY16 pKa = 10.54SFDD19 pKa = 3.58GRR21 pKa = 11.84IEE23 pKa = 3.84FANIDD28 pKa = 3.76NNIISHH34 pKa = 6.76PSPYY38 pKa = 10.31DD39 pKa = 3.21VEE41 pKa = 4.51FAANVALCSLPKK53 pKa = 10.46NEE55 pKa = 4.74EE56 pKa = 3.95GNVARR61 pKa = 11.84FDD63 pKa = 4.07YY64 pKa = 10.56SVHH67 pKa = 6.0PFEE70 pKa = 5.32IAVSGYY76 pKa = 9.96VDD78 pKa = 3.63GVFYY82 pKa = 10.02PASNPCMKK90 pKa = 10.16VFLLQVPCRR99 pKa = 11.84KK100 pKa = 9.87CEE102 pKa = 3.97LCCEE106 pKa = 4.18NRR108 pKa = 11.84KK109 pKa = 6.64QTYY112 pKa = 5.68VHH114 pKa = 7.2RR115 pKa = 11.84IMFEE119 pKa = 4.11AEE121 pKa = 3.91SSRR124 pKa = 11.84QVDD127 pKa = 4.34FVTLTFDD134 pKa = 3.58RR135 pKa = 11.84DD136 pKa = 3.72HH137 pKa = 6.59YY138 pKa = 10.01PSRR141 pKa = 11.84GATKK145 pKa = 10.35QEE147 pKa = 3.75LQRR150 pKa = 11.84FIDD153 pKa = 3.67SFRR156 pKa = 11.84KK157 pKa = 9.19FIKK160 pKa = 9.92RR161 pKa = 11.84YY162 pKa = 8.31SEE164 pKa = 3.8EE165 pKa = 3.93RR166 pKa = 11.84PRR168 pKa = 11.84TPKK171 pKa = 10.43VFDD174 pKa = 3.7GDD176 pKa = 3.25KK177 pKa = 10.15FRR179 pKa = 11.84YY180 pKa = 9.51IFVSEE185 pKa = 4.25YY186 pKa = 7.91GHH188 pKa = 6.92KK189 pKa = 10.81GRR191 pKa = 11.84GHH193 pKa = 5.1YY194 pKa = 9.78HH195 pKa = 6.96GEE197 pKa = 4.11MFNLPPYY204 pKa = 10.65EE205 pKa = 4.17EE206 pKa = 4.32FKK208 pKa = 10.91EE209 pKa = 4.66LYY211 pKa = 9.55ILQHH215 pKa = 5.56GTTLKK220 pKa = 11.08YY221 pKa = 9.62ITDD224 pKa = 3.97PQNSRR229 pKa = 11.84TVFYY233 pKa = 10.75EE234 pKa = 4.01DD235 pKa = 4.73LNPAPFYY242 pKa = 9.99KK243 pKa = 10.37LKK245 pKa = 10.88RR246 pKa = 11.84KK247 pKa = 8.83GCPVLEE253 pKa = 4.37PSVHH257 pKa = 7.04RR258 pKa = 11.84DD259 pKa = 3.39YY260 pKa = 11.47LYY262 pKa = 10.92LDD264 pKa = 3.54YY265 pKa = 10.92YY266 pKa = 9.49VRR268 pKa = 11.84KK269 pKa = 9.32IWNNGLTCSEE279 pKa = 4.24PTKK282 pKa = 10.89DD283 pKa = 4.25SAGSYY288 pKa = 7.9CAKK291 pKa = 10.44YY292 pKa = 9.89INKK295 pKa = 8.71PSEE298 pKa = 4.15MVPEE302 pKa = 4.94GCDD305 pKa = 2.92KK306 pKa = 11.29CFVLASNKK314 pKa = 10.17NGGIGSQYY322 pKa = 9.31CKK324 pKa = 9.88TVIRR328 pKa = 11.84EE329 pKa = 4.04FLKK332 pKa = 10.92KK333 pKa = 10.53NPTALVYY340 pKa = 10.43KK341 pKa = 9.39FRR343 pKa = 11.84PRR345 pKa = 11.84HH346 pKa = 5.32SSLIYY351 pKa = 9.03NIPIIGYY358 pKa = 8.4VKK360 pKa = 10.57NLALPRR366 pKa = 11.84PSDD369 pKa = 3.7VLSQRR374 pKa = 11.84YY375 pKa = 8.72VDD377 pKa = 3.71SFKK380 pKa = 10.54IYY382 pKa = 9.09HH383 pKa = 6.83WYY385 pKa = 10.38NRR387 pKa = 11.84MIGDD391 pKa = 3.45VLEE394 pKa = 4.24YY395 pKa = 10.65QFAEE399 pKa = 4.15AFFAPRR405 pKa = 11.84GCIKK409 pKa = 10.33PFRR412 pKa = 11.84STDD415 pKa = 3.37YY416 pKa = 10.34RR417 pKa = 11.84RR418 pKa = 11.84KK419 pKa = 10.55NPTEE423 pKa = 3.64YY424 pKa = 10.94EE425 pKa = 3.95KK426 pKa = 10.75FIKK429 pKa = 10.22KK430 pKa = 9.36NRR432 pKa = 11.84SKK434 pKa = 11.5EE435 pKa = 4.11EE436 pKa = 3.33ISKK439 pKa = 10.47IEE441 pKa = 4.28DD442 pKa = 3.18YY443 pKa = 11.2LYY445 pKa = 10.31IEE447 pKa = 4.78RR448 pKa = 11.84QNIDD452 pKa = 2.62WNEE455 pKa = 3.79IFKK458 pKa = 10.47LAHH461 pKa = 6.04LRR463 pKa = 11.84VINNKK468 pKa = 9.04AFRR471 pKa = 11.84DD472 pKa = 3.88SEE474 pKa = 3.98IPYY477 pKa = 9.91IDD479 pKa = 4.76DD480 pKa = 4.31LSDD483 pKa = 3.14EE484 pKa = 4.14LRR486 pKa = 11.84EE487 pKa = 4.05VRR489 pKa = 11.84SKK491 pKa = 11.26NLWRR495 pKa = 11.84KK496 pKa = 8.51EE497 pKa = 4.0RR498 pKa = 11.84IKK500 pKa = 11.06LL501 pKa = 3.62
MM1 pKa = 7.51KK2 pKa = 10.17CLNPVIMINRR12 pKa = 11.84PFLYY16 pKa = 10.54SFDD19 pKa = 3.58GRR21 pKa = 11.84IEE23 pKa = 3.84FANIDD28 pKa = 3.76NNIISHH34 pKa = 6.76PSPYY38 pKa = 10.31DD39 pKa = 3.21VEE41 pKa = 4.51FAANVALCSLPKK53 pKa = 10.46NEE55 pKa = 4.74EE56 pKa = 3.95GNVARR61 pKa = 11.84FDD63 pKa = 4.07YY64 pKa = 10.56SVHH67 pKa = 6.0PFEE70 pKa = 5.32IAVSGYY76 pKa = 9.96VDD78 pKa = 3.63GVFYY82 pKa = 10.02PASNPCMKK90 pKa = 10.16VFLLQVPCRR99 pKa = 11.84KK100 pKa = 9.87CEE102 pKa = 3.97LCCEE106 pKa = 4.18NRR108 pKa = 11.84KK109 pKa = 6.64QTYY112 pKa = 5.68VHH114 pKa = 7.2RR115 pKa = 11.84IMFEE119 pKa = 4.11AEE121 pKa = 3.91SSRR124 pKa = 11.84QVDD127 pKa = 4.34FVTLTFDD134 pKa = 3.58RR135 pKa = 11.84DD136 pKa = 3.72HH137 pKa = 6.59YY138 pKa = 10.01PSRR141 pKa = 11.84GATKK145 pKa = 10.35QEE147 pKa = 3.75LQRR150 pKa = 11.84FIDD153 pKa = 3.67SFRR156 pKa = 11.84KK157 pKa = 9.19FIKK160 pKa = 9.92RR161 pKa = 11.84YY162 pKa = 8.31SEE164 pKa = 3.8EE165 pKa = 3.93RR166 pKa = 11.84PRR168 pKa = 11.84TPKK171 pKa = 10.43VFDD174 pKa = 3.7GDD176 pKa = 3.25KK177 pKa = 10.15FRR179 pKa = 11.84YY180 pKa = 9.51IFVSEE185 pKa = 4.25YY186 pKa = 7.91GHH188 pKa = 6.92KK189 pKa = 10.81GRR191 pKa = 11.84GHH193 pKa = 5.1YY194 pKa = 9.78HH195 pKa = 6.96GEE197 pKa = 4.11MFNLPPYY204 pKa = 10.65EE205 pKa = 4.17EE206 pKa = 4.32FKK208 pKa = 10.91EE209 pKa = 4.66LYY211 pKa = 9.55ILQHH215 pKa = 5.56GTTLKK220 pKa = 11.08YY221 pKa = 9.62ITDD224 pKa = 3.97PQNSRR229 pKa = 11.84TVFYY233 pKa = 10.75EE234 pKa = 4.01DD235 pKa = 4.73LNPAPFYY242 pKa = 9.99KK243 pKa = 10.37LKK245 pKa = 10.88RR246 pKa = 11.84KK247 pKa = 8.83GCPVLEE253 pKa = 4.37PSVHH257 pKa = 7.04RR258 pKa = 11.84DD259 pKa = 3.39YY260 pKa = 11.47LYY262 pKa = 10.92LDD264 pKa = 3.54YY265 pKa = 10.92YY266 pKa = 9.49VRR268 pKa = 11.84KK269 pKa = 9.32IWNNGLTCSEE279 pKa = 4.24PTKK282 pKa = 10.89DD283 pKa = 4.25SAGSYY288 pKa = 7.9CAKK291 pKa = 10.44YY292 pKa = 9.89INKK295 pKa = 8.71PSEE298 pKa = 4.15MVPEE302 pKa = 4.94GCDD305 pKa = 2.92KK306 pKa = 11.29CFVLASNKK314 pKa = 10.17NGGIGSQYY322 pKa = 9.31CKK324 pKa = 9.88TVIRR328 pKa = 11.84EE329 pKa = 4.04FLKK332 pKa = 10.92KK333 pKa = 10.53NPTALVYY340 pKa = 10.43KK341 pKa = 9.39FRR343 pKa = 11.84PRR345 pKa = 11.84HH346 pKa = 5.32SSLIYY351 pKa = 9.03NIPIIGYY358 pKa = 8.4VKK360 pKa = 10.57NLALPRR366 pKa = 11.84PSDD369 pKa = 3.7VLSQRR374 pKa = 11.84YY375 pKa = 8.72VDD377 pKa = 3.71SFKK380 pKa = 10.54IYY382 pKa = 9.09HH383 pKa = 6.83WYY385 pKa = 10.38NRR387 pKa = 11.84MIGDD391 pKa = 3.45VLEE394 pKa = 4.24YY395 pKa = 10.65QFAEE399 pKa = 4.15AFFAPRR405 pKa = 11.84GCIKK409 pKa = 10.33PFRR412 pKa = 11.84STDD415 pKa = 3.37YY416 pKa = 10.34RR417 pKa = 11.84RR418 pKa = 11.84KK419 pKa = 10.55NPTEE423 pKa = 3.64YY424 pKa = 10.94EE425 pKa = 3.95KK426 pKa = 10.75FIKK429 pKa = 10.22KK430 pKa = 9.36NRR432 pKa = 11.84SKK434 pKa = 11.5EE435 pKa = 4.11EE436 pKa = 3.33ISKK439 pKa = 10.47IEE441 pKa = 4.28DD442 pKa = 3.18YY443 pKa = 11.2LYY445 pKa = 10.31IEE447 pKa = 4.78RR448 pKa = 11.84QNIDD452 pKa = 2.62WNEE455 pKa = 3.79IFKK458 pKa = 10.47LAHH461 pKa = 6.04LRR463 pKa = 11.84VINNKK468 pKa = 9.04AFRR471 pKa = 11.84DD472 pKa = 3.88SEE474 pKa = 3.98IPYY477 pKa = 9.91IDD479 pKa = 4.76DD480 pKa = 4.31LSDD483 pKa = 3.14EE484 pKa = 4.14LRR486 pKa = 11.84EE487 pKa = 4.05VRR489 pKa = 11.84SKK491 pKa = 11.26NLWRR495 pKa = 11.84KK496 pKa = 8.51EE497 pKa = 4.0RR498 pKa = 11.84IKK500 pKa = 11.06LL501 pKa = 3.62
Molecular weight: 59.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1777 |
72 |
738 |
355.4 |
40.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.684 ± 1.18 | 1.463 ± 0.538 |
5.796 ± 0.476 | 6.134 ± 1.03 |
5.008 ± 0.858 | 5.459 ± 0.77 |
1.745 ± 0.324 | 6.19 ± 0.754 |
5.965 ± 0.785 | 7.485 ± 0.879 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.364 ± 0.597 | 6.922 ± 1.012 |
4.389 ± 0.721 | 4.615 ± 1.435 |
5.29 ± 1.091 | 7.372 ± 0.915 |
5.402 ± 0.996 | 6.415 ± 1.394 |
1.182 ± 0.193 | 5.121 ± 1.128 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
