
Nitratiruptor sp. (strain SB155-2)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Epsilonproteobacteria; Nautiliales; Nitratiruptoraceae; Nitratiruptor; unclassified Nitratiruptor
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
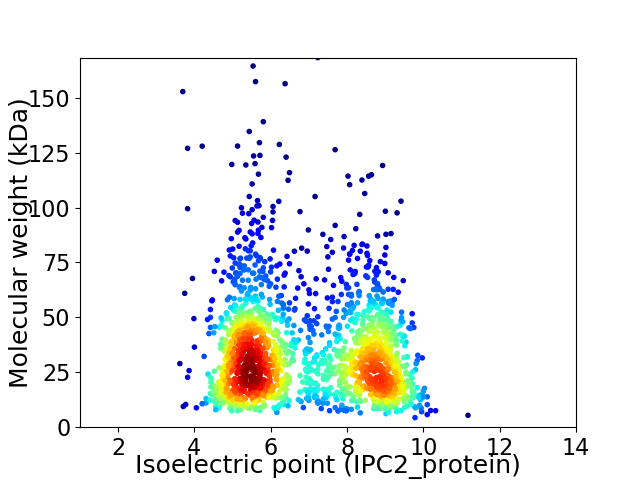
Virtual 2D-PAGE plot for 1843 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A6Q4W2|A6Q4W2_NITSB Polysaccharide biosynthesis protein OS=Nitratiruptor sp. (strain SB155-2) OX=387092 GN=NIS_1414 PE=4 SV=1
MM1 pKa = 7.48LTRR4 pKa = 11.84DD5 pKa = 4.29DD6 pKa = 3.91VAGLYY11 pKa = 9.62IALFQRR17 pKa = 11.84APSKK21 pKa = 11.16SEE23 pKa = 3.66LDD25 pKa = 2.82TWYY28 pKa = 11.25NDD30 pKa = 3.06VVANNRR36 pKa = 11.84DD37 pKa = 3.49LADD40 pKa = 3.5TAEE43 pKa = 4.3TMLQAAQLAVNGFGLQDD60 pKa = 5.12LYY62 pKa = 10.71PQYY65 pKa = 11.89ANVDD69 pKa = 3.53PSNPEE74 pKa = 3.66SVRR77 pKa = 11.84EE78 pKa = 4.13IISTVYY84 pKa = 8.37EE85 pKa = 3.94TLFNKK90 pKa = 9.55TYY92 pKa = 10.63QEE94 pKa = 4.48DD95 pKa = 4.04PQGVDD100 pKa = 2.35SWVDD104 pKa = 3.36LVVSGQQSLGEE115 pKa = 4.43AIAGITYY122 pKa = 9.73IGEE125 pKa = 4.72GIATKK130 pKa = 9.93PDD132 pKa = 3.12EE133 pKa = 4.26YY134 pKa = 10.82RR135 pKa = 11.84PYY137 pKa = 10.65FATDD141 pKa = 3.21DD142 pKa = 4.48DD143 pKa = 4.63FNKK146 pKa = 10.65AYY148 pKa = 10.31VAAKK152 pKa = 10.09AYY154 pKa = 7.72EE155 pKa = 4.15AKK157 pKa = 10.15KK158 pKa = 10.65DD159 pKa = 3.56AALEE163 pKa = 4.11VADD166 pKa = 4.47TIEE169 pKa = 4.05NVKK172 pKa = 10.04VDD174 pKa = 3.94KK175 pKa = 10.13DD176 pKa = 3.88TLQKK180 pKa = 8.92MQEE183 pKa = 4.04IIEE186 pKa = 4.3DD187 pKa = 3.92VKK189 pKa = 11.58DD190 pKa = 3.57EE191 pKa = 4.22TDD193 pKa = 3.38LDD195 pKa = 4.11EE196 pKa = 4.84VKK198 pKa = 11.0NIADD202 pKa = 3.66YY203 pKa = 11.35LKK205 pKa = 10.86DD206 pKa = 3.29IVEE209 pKa = 4.82DD210 pKa = 4.2PNTVSSNDD218 pKa = 3.69LNDD221 pKa = 3.57QNSSQIQNNLSDD233 pKa = 3.72DD234 pKa = 4.38TYY236 pKa = 11.75NDD238 pKa = 3.4AYY240 pKa = 10.65GYY242 pKa = 11.21NFDD245 pKa = 3.74TDD247 pKa = 3.33AWPSIMMEE255 pKa = 4.06LQMLPVEE262 pKa = 4.55GRR264 pKa = 11.84VEE266 pKa = 4.04LVNLMKK272 pKa = 10.64EE273 pKa = 4.21AVDD276 pKa = 4.24KK277 pKa = 11.13YY278 pKa = 11.39LSLAQANVPVTQEE291 pKa = 4.08YY292 pKa = 10.36LEE294 pKa = 4.0QWAALDD300 pKa = 3.99NEE302 pKa = 4.82FEE304 pKa = 4.49HH305 pKa = 6.78KK306 pKa = 10.42RR307 pKa = 11.84ADD309 pKa = 3.11IYY311 pKa = 11.03QKK313 pKa = 10.57HH314 pKa = 5.54GLPYY318 pKa = 10.31FIEE321 pKa = 4.26EE322 pKa = 4.36SVADD326 pKa = 4.2EE327 pKa = 4.62SFLNLDD333 pKa = 3.61PNIQRR338 pKa = 11.84TYY340 pKa = 10.05FQLEE344 pKa = 3.85KK345 pKa = 10.85DD346 pKa = 4.06YY347 pKa = 11.4LPKK350 pKa = 10.59LKK352 pKa = 10.48QEE354 pKa = 4.15LQYY357 pKa = 10.84VDD359 pKa = 4.09PMNVDD364 pKa = 2.81SFYY367 pKa = 11.03TLFKK371 pKa = 11.16LEE373 pKa = 4.72DD374 pKa = 3.99EE375 pKa = 4.49MTEE378 pKa = 4.15SQDD381 pKa = 3.39KK382 pKa = 9.02LTSGVGPTYY391 pKa = 10.38IPEE394 pKa = 4.21DD395 pKa = 3.35NYY397 pKa = 11.41YY398 pKa = 11.16DD399 pKa = 4.21FDD401 pKa = 3.82NTQNSMDD408 pKa = 4.14NYY410 pKa = 10.46YY411 pKa = 10.9NDD413 pKa = 3.62QISYY417 pKa = 11.14GDD419 pKa = 4.68LATADD424 pKa = 4.23NNALDD429 pKa = 4.24NDD431 pKa = 4.37LTTTTQTDD439 pKa = 3.62NYY441 pKa = 9.36NANAMSALEE450 pKa = 4.96DD451 pKa = 3.59MMASYY456 pKa = 10.2YY457 pKa = 10.72AQGGAYY463 pKa = 9.13CDD465 pKa = 4.0NIIASKK471 pKa = 9.08LTSTLSQDD479 pKa = 3.68QLVSLAKK486 pKa = 10.41DD487 pKa = 2.94IQNYY491 pKa = 8.95VFTSPYY497 pKa = 9.72FQDD500 pKa = 3.46GQITYY505 pKa = 10.8DD506 pKa = 3.61EE507 pKa = 4.43YY508 pKa = 11.64QPIANDD514 pKa = 3.82MIQIYY519 pKa = 10.08NKK521 pKa = 10.57YY522 pKa = 10.52GIDD525 pKa = 4.11LMDD528 pKa = 5.05LASDD532 pKa = 5.11CYY534 pKa = 11.43AIMM537 pKa = 5.07
MM1 pKa = 7.48LTRR4 pKa = 11.84DD5 pKa = 4.29DD6 pKa = 3.91VAGLYY11 pKa = 9.62IALFQRR17 pKa = 11.84APSKK21 pKa = 11.16SEE23 pKa = 3.66LDD25 pKa = 2.82TWYY28 pKa = 11.25NDD30 pKa = 3.06VVANNRR36 pKa = 11.84DD37 pKa = 3.49LADD40 pKa = 3.5TAEE43 pKa = 4.3TMLQAAQLAVNGFGLQDD60 pKa = 5.12LYY62 pKa = 10.71PQYY65 pKa = 11.89ANVDD69 pKa = 3.53PSNPEE74 pKa = 3.66SVRR77 pKa = 11.84EE78 pKa = 4.13IISTVYY84 pKa = 8.37EE85 pKa = 3.94TLFNKK90 pKa = 9.55TYY92 pKa = 10.63QEE94 pKa = 4.48DD95 pKa = 4.04PQGVDD100 pKa = 2.35SWVDD104 pKa = 3.36LVVSGQQSLGEE115 pKa = 4.43AIAGITYY122 pKa = 9.73IGEE125 pKa = 4.72GIATKK130 pKa = 9.93PDD132 pKa = 3.12EE133 pKa = 4.26YY134 pKa = 10.82RR135 pKa = 11.84PYY137 pKa = 10.65FATDD141 pKa = 3.21DD142 pKa = 4.48DD143 pKa = 4.63FNKK146 pKa = 10.65AYY148 pKa = 10.31VAAKK152 pKa = 10.09AYY154 pKa = 7.72EE155 pKa = 4.15AKK157 pKa = 10.15KK158 pKa = 10.65DD159 pKa = 3.56AALEE163 pKa = 4.11VADD166 pKa = 4.47TIEE169 pKa = 4.05NVKK172 pKa = 10.04VDD174 pKa = 3.94KK175 pKa = 10.13DD176 pKa = 3.88TLQKK180 pKa = 8.92MQEE183 pKa = 4.04IIEE186 pKa = 4.3DD187 pKa = 3.92VKK189 pKa = 11.58DD190 pKa = 3.57EE191 pKa = 4.22TDD193 pKa = 3.38LDD195 pKa = 4.11EE196 pKa = 4.84VKK198 pKa = 11.0NIADD202 pKa = 3.66YY203 pKa = 11.35LKK205 pKa = 10.86DD206 pKa = 3.29IVEE209 pKa = 4.82DD210 pKa = 4.2PNTVSSNDD218 pKa = 3.69LNDD221 pKa = 3.57QNSSQIQNNLSDD233 pKa = 3.72DD234 pKa = 4.38TYY236 pKa = 11.75NDD238 pKa = 3.4AYY240 pKa = 10.65GYY242 pKa = 11.21NFDD245 pKa = 3.74TDD247 pKa = 3.33AWPSIMMEE255 pKa = 4.06LQMLPVEE262 pKa = 4.55GRR264 pKa = 11.84VEE266 pKa = 4.04LVNLMKK272 pKa = 10.64EE273 pKa = 4.21AVDD276 pKa = 4.24KK277 pKa = 11.13YY278 pKa = 11.39LSLAQANVPVTQEE291 pKa = 4.08YY292 pKa = 10.36LEE294 pKa = 4.0QWAALDD300 pKa = 3.99NEE302 pKa = 4.82FEE304 pKa = 4.49HH305 pKa = 6.78KK306 pKa = 10.42RR307 pKa = 11.84ADD309 pKa = 3.11IYY311 pKa = 11.03QKK313 pKa = 10.57HH314 pKa = 5.54GLPYY318 pKa = 10.31FIEE321 pKa = 4.26EE322 pKa = 4.36SVADD326 pKa = 4.2EE327 pKa = 4.62SFLNLDD333 pKa = 3.61PNIQRR338 pKa = 11.84TYY340 pKa = 10.05FQLEE344 pKa = 3.85KK345 pKa = 10.85DD346 pKa = 4.06YY347 pKa = 11.4LPKK350 pKa = 10.59LKK352 pKa = 10.48QEE354 pKa = 4.15LQYY357 pKa = 10.84VDD359 pKa = 4.09PMNVDD364 pKa = 2.81SFYY367 pKa = 11.03TLFKK371 pKa = 11.16LEE373 pKa = 4.72DD374 pKa = 3.99EE375 pKa = 4.49MTEE378 pKa = 4.15SQDD381 pKa = 3.39KK382 pKa = 9.02LTSGVGPTYY391 pKa = 10.38IPEE394 pKa = 4.21DD395 pKa = 3.35NYY397 pKa = 11.41YY398 pKa = 11.16DD399 pKa = 4.21FDD401 pKa = 3.82NTQNSMDD408 pKa = 4.14NYY410 pKa = 10.46YY411 pKa = 10.9NDD413 pKa = 3.62QISYY417 pKa = 11.14GDD419 pKa = 4.68LATADD424 pKa = 4.23NNALDD429 pKa = 4.24NDD431 pKa = 4.37LTTTTQTDD439 pKa = 3.62NYY441 pKa = 9.36NANAMSALEE450 pKa = 4.96DD451 pKa = 3.59MMASYY456 pKa = 10.2YY457 pKa = 10.72AQGGAYY463 pKa = 9.13CDD465 pKa = 4.0NIIASKK471 pKa = 9.08LTSTLSQDD479 pKa = 3.68QLVSLAKK486 pKa = 10.41DD487 pKa = 2.94IQNYY491 pKa = 8.95VFTSPYY497 pKa = 9.72FQDD500 pKa = 3.46GQITYY505 pKa = 10.8DD506 pKa = 3.61EE507 pKa = 4.43YY508 pKa = 11.64QPIANDD514 pKa = 3.82MIQIYY519 pKa = 10.08NKK521 pKa = 10.57YY522 pKa = 10.52GIDD525 pKa = 4.11LMDD528 pKa = 5.05LASDD532 pKa = 5.11CYY534 pKa = 11.43AIMM537 pKa = 5.07
Molecular weight: 60.96 kDa
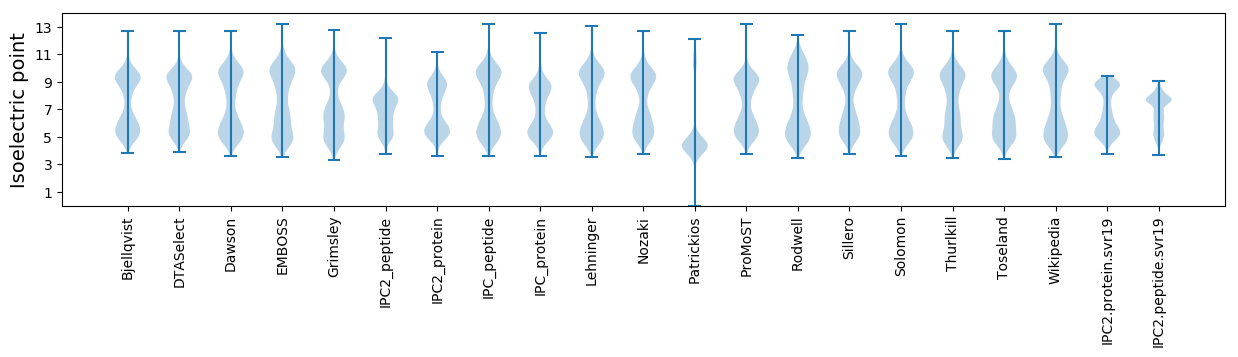
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A6Q3D3|A6Q3D3_NITSB Putative membrane protein insertion efficiency factor OS=Nitratiruptor sp. (strain SB155-2) OX=387092 GN=NIS_0880 PE=3 SV=1
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.44NTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 7.5RR14 pKa = 11.84THH16 pKa = 5.97GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.78TKK25 pKa = 10.46NGRR28 pKa = 11.84KK29 pKa = 9.26VINARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.61GRR39 pKa = 11.84KK40 pKa = 8.79RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.04QPHH8 pKa = 4.44NTRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 7.5RR14 pKa = 11.84THH16 pKa = 5.97GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.78TKK25 pKa = 10.46NGRR28 pKa = 11.84KK29 pKa = 9.26VINARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.61GRR39 pKa = 11.84KK40 pKa = 8.79RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.42 kDa
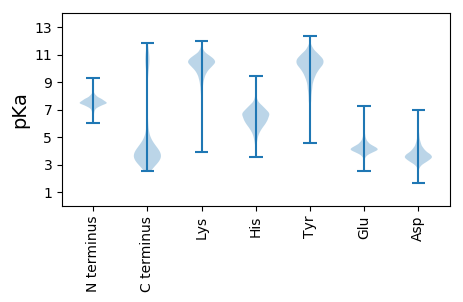
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
587077 |
37 |
1507 |
318.5 |
36.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.983 ± 0.058 | 0.875 ± 0.021 |
5.325 ± 0.046 | 7.473 ± 0.071 |
5.144 ± 0.053 | 5.968 ± 0.052 |
2.07 ± 0.025 | 8.501 ± 0.057 |
8.914 ± 0.067 | 9.936 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.356 ± 0.03 | 4.252 ± 0.046 |
3.589 ± 0.032 | 3.375 ± 0.032 |
3.968 ± 0.035 | 5.426 ± 0.043 |
4.633 ± 0.041 | 6.285 ± 0.042 |
0.858 ± 0.021 | 4.069 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
