
Hirame rhabdovirus (strain Korea/CA 9703/1997) (HIRRV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Gammarhabdovirinae; Novirhabdovirus; Hirame novirhabdovirus
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
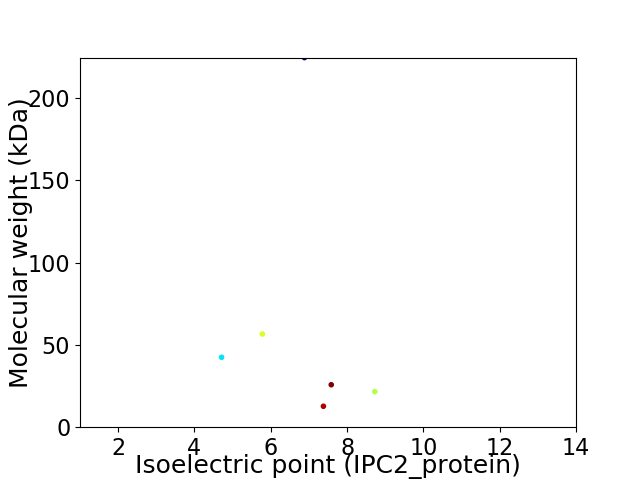
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q9QL89|NCAP_HIRRV Nucleoprotein OS=Hirame rhabdovirus (strain Korea/CA 9703/1997) OX=453457 GN=N PE=3 SV=1
MM1 pKa = 7.71ANLKK5 pKa = 10.57EE6 pKa = 4.19EE7 pKa = 4.3FAGLRR12 pKa = 11.84GVKK15 pKa = 10.14GGALEE20 pKa = 4.92DD21 pKa = 3.86SGTEE25 pKa = 3.88YY26 pKa = 11.28DD27 pKa = 3.56PTKK30 pKa = 10.77INLTLYY36 pKa = 9.17GTDD39 pKa = 3.24KK40 pKa = 11.1LYY42 pKa = 10.24TLAIIKK48 pKa = 10.17RR49 pKa = 11.84AVSQVGGSQTNKK61 pKa = 10.74ALGILCAFVTSEE73 pKa = 4.54NNPDD77 pKa = 3.26MTDD80 pKa = 3.05AAVKK84 pKa = 10.33LLVDD88 pKa = 3.48MRR90 pKa = 11.84FKK92 pKa = 11.25VDD94 pKa = 3.42VVPVDD99 pKa = 4.87DD100 pKa = 5.17RR101 pKa = 11.84LGDD104 pKa = 3.83NLDD107 pKa = 4.36DD108 pKa = 5.08PNSKK112 pKa = 10.06LAEE115 pKa = 4.16VLTEE119 pKa = 3.94EE120 pKa = 5.06NMVDD124 pKa = 3.75LVKK127 pKa = 11.04GLLFTCALMVKK138 pKa = 10.31YY139 pKa = 10.39DD140 pKa = 3.7VDD142 pKa = 4.63KK143 pKa = 10.85MATYY147 pKa = 9.84CQQKK151 pKa = 10.36LEE153 pKa = 4.33RR154 pKa = 11.84LANSQGLNEE163 pKa = 4.5LTLISTSRR171 pKa = 11.84AVLARR176 pKa = 11.84IGAAVRR182 pKa = 11.84PGQKK186 pKa = 8.37LTKK189 pKa = 9.73AIYY192 pKa = 9.93GIILINLMDD201 pKa = 5.24PATAARR207 pKa = 11.84AKK209 pKa = 9.97ALCAMRR215 pKa = 11.84LSGTGMTMVGLFNQASKK232 pKa = 11.41NLGAPPADD240 pKa = 3.93LLEE243 pKa = 4.49DD244 pKa = 3.68LCMKK248 pKa = 10.58SIIDD252 pKa = 3.46SARR255 pKa = 11.84RR256 pKa = 11.84IVKK259 pKa = 10.06LMRR262 pKa = 11.84IVADD266 pKa = 3.9VEE268 pKa = 4.88DD269 pKa = 4.14MTAKK273 pKa = 9.91YY274 pKa = 10.47AIMMSRR280 pKa = 11.84MLGDD284 pKa = 4.73GYY286 pKa = 10.94FKK288 pKa = 10.93SYY290 pKa = 10.79GINEE294 pKa = 4.2NSRR297 pKa = 11.84ITCILMNINEE307 pKa = 4.38QYY309 pKa = 11.33DD310 pKa = 3.43EE311 pKa = 4.33GTTGGLAGVRR321 pKa = 11.84VSPPFRR327 pKa = 11.84KK328 pKa = 9.57LATEE332 pKa = 4.32IARR335 pKa = 11.84LLVKK339 pKa = 10.38KK340 pKa = 10.82YY341 pKa = 10.44DD342 pKa = 4.05GNGSAGPGASDD353 pKa = 4.26LVRR356 pKa = 11.84QAEE359 pKa = 4.04QAAQEE364 pKa = 4.47TEE366 pKa = 4.12GEE368 pKa = 3.89DD369 pKa = 3.75DD370 pKa = 4.68KK371 pKa = 11.86YY372 pKa = 11.52DD373 pKa = 3.71EE374 pKa = 4.96EE375 pKa = 5.78GEE377 pKa = 4.04EE378 pKa = 4.66DD379 pKa = 3.51GGEE382 pKa = 4.34GEE384 pKa = 4.27GGEE387 pKa = 4.19DD388 pKa = 3.12EE389 pKa = 5.12EE390 pKa = 5.67YY391 pKa = 10.98YY392 pKa = 11.26
MM1 pKa = 7.71ANLKK5 pKa = 10.57EE6 pKa = 4.19EE7 pKa = 4.3FAGLRR12 pKa = 11.84GVKK15 pKa = 10.14GGALEE20 pKa = 4.92DD21 pKa = 3.86SGTEE25 pKa = 3.88YY26 pKa = 11.28DD27 pKa = 3.56PTKK30 pKa = 10.77INLTLYY36 pKa = 9.17GTDD39 pKa = 3.24KK40 pKa = 11.1LYY42 pKa = 10.24TLAIIKK48 pKa = 10.17RR49 pKa = 11.84AVSQVGGSQTNKK61 pKa = 10.74ALGILCAFVTSEE73 pKa = 4.54NNPDD77 pKa = 3.26MTDD80 pKa = 3.05AAVKK84 pKa = 10.33LLVDD88 pKa = 3.48MRR90 pKa = 11.84FKK92 pKa = 11.25VDD94 pKa = 3.42VVPVDD99 pKa = 4.87DD100 pKa = 5.17RR101 pKa = 11.84LGDD104 pKa = 3.83NLDD107 pKa = 4.36DD108 pKa = 5.08PNSKK112 pKa = 10.06LAEE115 pKa = 4.16VLTEE119 pKa = 3.94EE120 pKa = 5.06NMVDD124 pKa = 3.75LVKK127 pKa = 11.04GLLFTCALMVKK138 pKa = 10.31YY139 pKa = 10.39DD140 pKa = 3.7VDD142 pKa = 4.63KK143 pKa = 10.85MATYY147 pKa = 9.84CQQKK151 pKa = 10.36LEE153 pKa = 4.33RR154 pKa = 11.84LANSQGLNEE163 pKa = 4.5LTLISTSRR171 pKa = 11.84AVLARR176 pKa = 11.84IGAAVRR182 pKa = 11.84PGQKK186 pKa = 8.37LTKK189 pKa = 9.73AIYY192 pKa = 9.93GIILINLMDD201 pKa = 5.24PATAARR207 pKa = 11.84AKK209 pKa = 9.97ALCAMRR215 pKa = 11.84LSGTGMTMVGLFNQASKK232 pKa = 11.41NLGAPPADD240 pKa = 3.93LLEE243 pKa = 4.49DD244 pKa = 3.68LCMKK248 pKa = 10.58SIIDD252 pKa = 3.46SARR255 pKa = 11.84RR256 pKa = 11.84IVKK259 pKa = 10.06LMRR262 pKa = 11.84IVADD266 pKa = 3.9VEE268 pKa = 4.88DD269 pKa = 4.14MTAKK273 pKa = 9.91YY274 pKa = 10.47AIMMSRR280 pKa = 11.84MLGDD284 pKa = 4.73GYY286 pKa = 10.94FKK288 pKa = 10.93SYY290 pKa = 10.79GINEE294 pKa = 4.2NSRR297 pKa = 11.84ITCILMNINEE307 pKa = 4.38QYY309 pKa = 11.33DD310 pKa = 3.43EE311 pKa = 4.33GTTGGLAGVRR321 pKa = 11.84VSPPFRR327 pKa = 11.84KK328 pKa = 9.57LATEE332 pKa = 4.32IARR335 pKa = 11.84LLVKK339 pKa = 10.38KK340 pKa = 10.82YY341 pKa = 10.44DD342 pKa = 4.05GNGSAGPGASDD353 pKa = 4.26LVRR356 pKa = 11.84QAEE359 pKa = 4.04QAAQEE364 pKa = 4.47TEE366 pKa = 4.12GEE368 pKa = 3.89DD369 pKa = 3.75DD370 pKa = 4.68KK371 pKa = 11.86YY372 pKa = 11.52DD373 pKa = 3.71EE374 pKa = 4.96EE375 pKa = 5.78GEE377 pKa = 4.04EE378 pKa = 4.66DD379 pKa = 3.51GGEE382 pKa = 4.34GEE384 pKa = 4.27GGEE387 pKa = 4.19DD388 pKa = 3.12EE389 pKa = 5.12EE390 pKa = 5.67YY391 pKa = 10.98YY392 pKa = 11.26
Molecular weight: 42.47 kDa
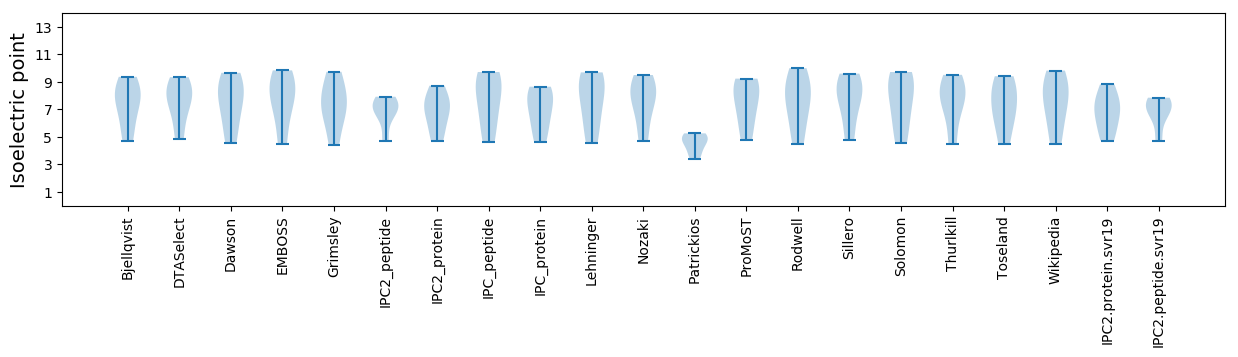
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q82080|NV_HIRRV Non-virion protein OS=Hirame rhabdovirus (strain Korea/CA 9703/1997) OX=453457 GN=NV PE=3 SV=1
MM1 pKa = 7.42SLFKK5 pKa = 9.97RR6 pKa = 11.84TKK8 pKa = 8.77KK9 pKa = 9.29TILIPPPHH17 pKa = 6.93LLSGDD22 pKa = 3.66EE23 pKa = 4.58DD24 pKa = 4.01RR25 pKa = 11.84VTVLNAEE32 pKa = 4.3GEE34 pKa = 4.33IKK36 pKa = 10.39ISGKK40 pKa = 10.33RR41 pKa = 11.84PTTLDD46 pKa = 3.1EE47 pKa = 4.65KK48 pKa = 10.5IYY50 pKa = 11.15YY51 pKa = 10.56SMSLAAAILGGNLHH65 pKa = 7.13PSFQSLTYY73 pKa = 10.69LFQQEE78 pKa = 4.49MEE80 pKa = 4.35FGSTIEE86 pKa = 4.06KK87 pKa = 10.78VNFGSRR93 pKa = 11.84KK94 pKa = 8.6PAPLTPIRR102 pKa = 11.84WQKK105 pKa = 10.16PEE107 pKa = 3.8KK108 pKa = 10.36CSFRR112 pKa = 11.84PQPLDD117 pKa = 3.51KK118 pKa = 10.59KK119 pKa = 10.35IPPQIYY125 pKa = 7.77TVSVEE130 pKa = 3.92GATITFSGRR139 pKa = 11.84FLFSASHH146 pKa = 6.24VGCDD150 pKa = 3.62DD151 pKa = 3.8NRR153 pKa = 11.84VKK155 pKa = 10.89LAGLDD160 pKa = 3.79GFITSPNYY168 pKa = 10.12QRR170 pKa = 11.84VKK172 pKa = 10.61DD173 pKa = 4.04YY174 pKa = 10.97YY175 pKa = 10.66AQEE178 pKa = 4.14TVLALSFEE186 pKa = 4.37IPTKK190 pKa = 10.29KK191 pKa = 10.58GKK193 pKa = 9.99
MM1 pKa = 7.42SLFKK5 pKa = 9.97RR6 pKa = 11.84TKK8 pKa = 8.77KK9 pKa = 9.29TILIPPPHH17 pKa = 6.93LLSGDD22 pKa = 3.66EE23 pKa = 4.58DD24 pKa = 4.01RR25 pKa = 11.84VTVLNAEE32 pKa = 4.3GEE34 pKa = 4.33IKK36 pKa = 10.39ISGKK40 pKa = 10.33RR41 pKa = 11.84PTTLDD46 pKa = 3.1EE47 pKa = 4.65KK48 pKa = 10.5IYY50 pKa = 11.15YY51 pKa = 10.56SMSLAAAILGGNLHH65 pKa = 7.13PSFQSLTYY73 pKa = 10.69LFQQEE78 pKa = 4.49MEE80 pKa = 4.35FGSTIEE86 pKa = 4.06KK87 pKa = 10.78VNFGSRR93 pKa = 11.84KK94 pKa = 8.6PAPLTPIRR102 pKa = 11.84WQKK105 pKa = 10.16PEE107 pKa = 3.8KK108 pKa = 10.36CSFRR112 pKa = 11.84PQPLDD117 pKa = 3.51KK118 pKa = 10.59KK119 pKa = 10.35IPPQIYY125 pKa = 7.77TVSVEE130 pKa = 3.92GATITFSGRR139 pKa = 11.84FLFSASHH146 pKa = 6.24VGCDD150 pKa = 3.62DD151 pKa = 3.8NRR153 pKa = 11.84VKK155 pKa = 10.89LAGLDD160 pKa = 3.79GFITSPNYY168 pKa = 10.12QRR170 pKa = 11.84VKK172 pKa = 10.61DD173 pKa = 4.04YY174 pKa = 10.97YY175 pKa = 10.66AQEE178 pKa = 4.14TVLALSFEE186 pKa = 4.37IPTKK190 pKa = 10.29KK191 pKa = 10.58GKK193 pKa = 9.99
Molecular weight: 21.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3417 |
111 |
1986 |
569.5 |
63.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.053 ± 0.625 | 1.405 ± 0.275 |
6.263 ± 0.457 | 6.555 ± 0.794 |
3.629 ± 0.424 | 6.292 ± 0.746 |
2.195 ± 0.54 | 5.999 ± 0.726 |
6.204 ± 0.469 | 11.062 ± 0.952 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.488 ± 0.466 | 3.161 ± 0.381 |
4.565 ± 0.596 | 3.395 ± 0.348 |
6.263 ± 0.548 | 7.112 ± 0.566 |
6.702 ± 0.588 | 5.531 ± 0.632 |
1.258 ± 0.36 | 2.868 ± 0.538 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
