
Bark beetle-associated genomovirus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus denpo1
Average proteome isoelectric point is 7.53
Get precalculated fractions of proteins
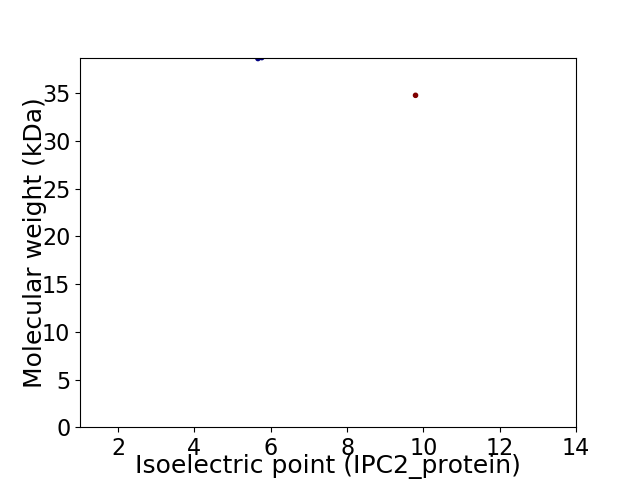
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A344A3Q5|A0A344A3Q5_9VIRU Replication-associated protein OS=Bark beetle-associated genomovirus 5 OX=2230900 PE=3 SV=1
MM1 pKa = 7.4LFVNSRR7 pKa = 11.84YY8 pKa = 10.23VLLTYY13 pKa = 10.38SQSNGLDD20 pKa = 2.94EE21 pKa = 4.5WAVSDD26 pKa = 4.22HH27 pKa = 6.04MSGLGAEE34 pKa = 4.59CIVARR39 pKa = 11.84EE40 pKa = 3.91NHH42 pKa = 6.36IDD44 pKa = 3.89GGTHH48 pKa = 5.55LHH50 pKa = 6.67AFVDD54 pKa = 4.91FNRR57 pKa = 11.84KK58 pKa = 8.42FRR60 pKa = 11.84SRR62 pKa = 11.84NARR65 pKa = 11.84VFDD68 pKa = 3.41VGGFHH73 pKa = 7.32PNVSPSRR80 pKa = 11.84GTPEE84 pKa = 3.81KK85 pKa = 10.76GYY87 pKa = 10.9DD88 pKa = 3.44YY89 pKa = 10.16ATKK92 pKa = 10.73DD93 pKa = 3.25GDD95 pKa = 3.98VVAGGLEE102 pKa = 4.02RR103 pKa = 11.84PAPRR107 pKa = 11.84GGLHH111 pKa = 6.16TGAHH115 pKa = 5.77RR116 pKa = 11.84VRR118 pKa = 11.84NVAHH122 pKa = 6.66LCEE125 pKa = 4.54SVDD128 pKa = 4.02EE129 pKa = 4.46FLEE132 pKa = 5.14LYY134 pKa = 10.95DD135 pKa = 3.62EE136 pKa = 4.52MEE138 pKa = 4.76RR139 pKa = 11.84GDD141 pKa = 4.1LIGRR145 pKa = 11.84FSNIRR150 pKa = 11.84AYY152 pKa = 10.57ADD154 pKa = 2.67WRR156 pKa = 11.84FRR158 pKa = 11.84VEE160 pKa = 3.89PPAYY164 pKa = 9.91SSPPGVDD171 pKa = 3.95FGSGAMDD178 pKa = 5.32GRR180 pKa = 11.84DD181 pKa = 3.29DD182 pKa = 4.53WISQSGICSGEE193 pKa = 3.81PLIGPRR199 pKa = 11.84RR200 pKa = 11.84KK201 pKa = 10.12SLVLFGPSLTGKK213 pKa = 5.84TTWARR218 pKa = 11.84SLGNHH223 pKa = 6.96IYY225 pKa = 8.65SQRR228 pKa = 11.84MLNVRR233 pKa = 11.84EE234 pKa = 3.86VAYY237 pKa = 10.87SIDD240 pKa = 3.29SARR243 pKa = 11.84YY244 pKa = 8.42HH245 pKa = 6.61VLDD248 pKa = 5.09DD249 pKa = 3.23VDD251 pKa = 4.78LRR253 pKa = 11.84YY254 pKa = 9.85FPGWKK259 pKa = 9.58DD260 pKa = 3.09WLGGMMWIPVRR271 pKa = 11.84LMYY274 pKa = 10.29RR275 pKa = 11.84DD276 pKa = 3.7VMNMQWGRR284 pKa = 11.84PCIWCNNTDD293 pKa = 3.35PRR295 pKa = 11.84DD296 pKa = 3.77VIKK299 pKa = 10.79RR300 pKa = 11.84SMAKK304 pKa = 9.87HH305 pKa = 6.7DD306 pKa = 3.9GQGDD310 pKa = 3.98GTFSQEE316 pKa = 4.08DD317 pKa = 4.44LDD319 pKa = 4.02WLEE322 pKa = 4.46ANCIFIRR329 pKa = 11.84VNDD332 pKa = 4.81PIVTFRR338 pKa = 11.84ASTEE342 pKa = 3.77
MM1 pKa = 7.4LFVNSRR7 pKa = 11.84YY8 pKa = 10.23VLLTYY13 pKa = 10.38SQSNGLDD20 pKa = 2.94EE21 pKa = 4.5WAVSDD26 pKa = 4.22HH27 pKa = 6.04MSGLGAEE34 pKa = 4.59CIVARR39 pKa = 11.84EE40 pKa = 3.91NHH42 pKa = 6.36IDD44 pKa = 3.89GGTHH48 pKa = 5.55LHH50 pKa = 6.67AFVDD54 pKa = 4.91FNRR57 pKa = 11.84KK58 pKa = 8.42FRR60 pKa = 11.84SRR62 pKa = 11.84NARR65 pKa = 11.84VFDD68 pKa = 3.41VGGFHH73 pKa = 7.32PNVSPSRR80 pKa = 11.84GTPEE84 pKa = 3.81KK85 pKa = 10.76GYY87 pKa = 10.9DD88 pKa = 3.44YY89 pKa = 10.16ATKK92 pKa = 10.73DD93 pKa = 3.25GDD95 pKa = 3.98VVAGGLEE102 pKa = 4.02RR103 pKa = 11.84PAPRR107 pKa = 11.84GGLHH111 pKa = 6.16TGAHH115 pKa = 5.77RR116 pKa = 11.84VRR118 pKa = 11.84NVAHH122 pKa = 6.66LCEE125 pKa = 4.54SVDD128 pKa = 4.02EE129 pKa = 4.46FLEE132 pKa = 5.14LYY134 pKa = 10.95DD135 pKa = 3.62EE136 pKa = 4.52MEE138 pKa = 4.76RR139 pKa = 11.84GDD141 pKa = 4.1LIGRR145 pKa = 11.84FSNIRR150 pKa = 11.84AYY152 pKa = 10.57ADD154 pKa = 2.67WRR156 pKa = 11.84FRR158 pKa = 11.84VEE160 pKa = 3.89PPAYY164 pKa = 9.91SSPPGVDD171 pKa = 3.95FGSGAMDD178 pKa = 5.32GRR180 pKa = 11.84DD181 pKa = 3.29DD182 pKa = 4.53WISQSGICSGEE193 pKa = 3.81PLIGPRR199 pKa = 11.84RR200 pKa = 11.84KK201 pKa = 10.12SLVLFGPSLTGKK213 pKa = 5.84TTWARR218 pKa = 11.84SLGNHH223 pKa = 6.96IYY225 pKa = 8.65SQRR228 pKa = 11.84MLNVRR233 pKa = 11.84EE234 pKa = 3.86VAYY237 pKa = 10.87SIDD240 pKa = 3.29SARR243 pKa = 11.84YY244 pKa = 8.42HH245 pKa = 6.61VLDD248 pKa = 5.09DD249 pKa = 3.23VDD251 pKa = 4.78LRR253 pKa = 11.84YY254 pKa = 9.85FPGWKK259 pKa = 9.58DD260 pKa = 3.09WLGGMMWIPVRR271 pKa = 11.84LMYY274 pKa = 10.29RR275 pKa = 11.84DD276 pKa = 3.7VMNMQWGRR284 pKa = 11.84PCIWCNNTDD293 pKa = 3.35PRR295 pKa = 11.84DD296 pKa = 3.77VIKK299 pKa = 10.79RR300 pKa = 11.84SMAKK304 pKa = 9.87HH305 pKa = 6.7DD306 pKa = 3.9GQGDD310 pKa = 3.98GTFSQEE316 pKa = 4.08DD317 pKa = 4.44LDD319 pKa = 4.02WLEE322 pKa = 4.46ANCIFIRR329 pKa = 11.84VNDD332 pKa = 4.81PIVTFRR338 pKa = 11.84ASTEE342 pKa = 3.77
Molecular weight: 38.66 kDa
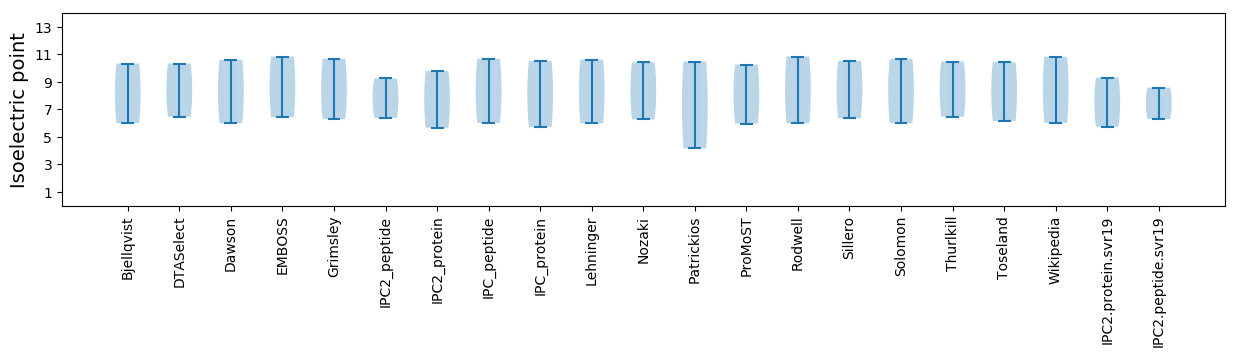
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A344A3Q5|A0A344A3Q5_9VIRU Replication-associated protein OS=Bark beetle-associated genomovirus 5 OX=2230900 PE=3 SV=1
MM1 pKa = 7.57SYY3 pKa = 10.55SSKK6 pKa = 10.38SRR8 pKa = 11.84SVRR11 pKa = 11.84VSRR14 pKa = 11.84RR15 pKa = 11.84FKK17 pKa = 10.23SRR19 pKa = 11.84NRR21 pKa = 11.84HH22 pKa = 5.01RR23 pKa = 11.84GSRR26 pKa = 11.84RR27 pKa = 11.84SKK29 pKa = 8.11RR30 pKa = 11.84TNRR33 pKa = 11.84RR34 pKa = 11.84SARR37 pKa = 11.84STYY40 pKa = 10.07RR41 pKa = 11.84KK42 pKa = 10.22GMTRR46 pKa = 11.84KK47 pKa = 9.9RR48 pKa = 11.84ILNITSRR55 pKa = 11.84KK56 pKa = 8.35KK57 pKa = 10.37RR58 pKa = 11.84NGMLCVTNSGTTGVGTSTFTNGALYY83 pKa = 11.2VNGSSGGWVLWAATAQDD100 pKa = 4.62LNDD103 pKa = 3.93GSSTLGTVAEE113 pKa = 4.42MAQRR117 pKa = 11.84TASTCYY123 pKa = 8.93MRR125 pKa = 11.84GLSEE129 pKa = 4.66HH130 pKa = 6.85IKK132 pKa = 10.07IQTSSAIPWFWRR144 pKa = 11.84RR145 pKa = 11.84IVFRR149 pKa = 11.84LKK151 pKa = 10.91GNPTFQAYY159 pKa = 8.12TSAPPNLTGPYY170 pKa = 8.56YY171 pKa = 10.8DD172 pKa = 3.37STAGLEE178 pKa = 3.93RR179 pKa = 11.84LLLNSNSQTVSQSGQVSAQQSVLFKK204 pKa = 10.92GRR206 pKa = 11.84AGFDD210 pKa = 3.16WNDD213 pKa = 3.53PLIAPVDD220 pKa = 3.73TARR223 pKa = 11.84VSLMYY228 pKa = 10.72DD229 pKa = 2.88KK230 pKa = 11.04TRR232 pKa = 11.84TLRR235 pKa = 11.84SGNQSGALLEE245 pKa = 4.58SKK247 pKa = 10.37LFHH250 pKa = 6.72SFNKK254 pKa = 9.98NLVYY258 pKa = 10.88DD259 pKa = 4.57DD260 pKa = 5.2DD261 pKa = 4.72EE262 pKa = 6.51SGATEE267 pKa = 3.85EE268 pKa = 3.89TSYY271 pKa = 11.4FSVQSKK277 pKa = 11.0AGMGDD282 pKa = 3.78VYY284 pKa = 11.56VLDD287 pKa = 3.55QMYY290 pKa = 10.74AGAGASATDD299 pKa = 4.13LLQFSTSSTLYY310 pKa = 7.41WHH312 pKa = 6.65EE313 pKa = 4.0RR314 pKa = 3.41
MM1 pKa = 7.57SYY3 pKa = 10.55SSKK6 pKa = 10.38SRR8 pKa = 11.84SVRR11 pKa = 11.84VSRR14 pKa = 11.84RR15 pKa = 11.84FKK17 pKa = 10.23SRR19 pKa = 11.84NRR21 pKa = 11.84HH22 pKa = 5.01RR23 pKa = 11.84GSRR26 pKa = 11.84RR27 pKa = 11.84SKK29 pKa = 8.11RR30 pKa = 11.84TNRR33 pKa = 11.84RR34 pKa = 11.84SARR37 pKa = 11.84STYY40 pKa = 10.07RR41 pKa = 11.84KK42 pKa = 10.22GMTRR46 pKa = 11.84KK47 pKa = 9.9RR48 pKa = 11.84ILNITSRR55 pKa = 11.84KK56 pKa = 8.35KK57 pKa = 10.37RR58 pKa = 11.84NGMLCVTNSGTTGVGTSTFTNGALYY83 pKa = 11.2VNGSSGGWVLWAATAQDD100 pKa = 4.62LNDD103 pKa = 3.93GSSTLGTVAEE113 pKa = 4.42MAQRR117 pKa = 11.84TASTCYY123 pKa = 8.93MRR125 pKa = 11.84GLSEE129 pKa = 4.66HH130 pKa = 6.85IKK132 pKa = 10.07IQTSSAIPWFWRR144 pKa = 11.84RR145 pKa = 11.84IVFRR149 pKa = 11.84LKK151 pKa = 10.91GNPTFQAYY159 pKa = 8.12TSAPPNLTGPYY170 pKa = 8.56YY171 pKa = 10.8DD172 pKa = 3.37STAGLEE178 pKa = 3.93RR179 pKa = 11.84LLLNSNSQTVSQSGQVSAQQSVLFKK204 pKa = 10.92GRR206 pKa = 11.84AGFDD210 pKa = 3.16WNDD213 pKa = 3.53PLIAPVDD220 pKa = 3.73TARR223 pKa = 11.84VSLMYY228 pKa = 10.72DD229 pKa = 2.88KK230 pKa = 11.04TRR232 pKa = 11.84TLRR235 pKa = 11.84SGNQSGALLEE245 pKa = 4.58SKK247 pKa = 10.37LFHH250 pKa = 6.72SFNKK254 pKa = 9.98NLVYY258 pKa = 10.88DD259 pKa = 4.57DD260 pKa = 5.2DD261 pKa = 4.72EE262 pKa = 6.51SGATEE267 pKa = 3.85EE268 pKa = 3.89TSYY271 pKa = 11.4FSVQSKK277 pKa = 11.0AGMGDD282 pKa = 3.78VYY284 pKa = 11.56VLDD287 pKa = 3.55QMYY290 pKa = 10.74AGAGASATDD299 pKa = 4.13LLQFSTSSTLYY310 pKa = 7.41WHH312 pKa = 6.65EE313 pKa = 4.0RR314 pKa = 3.41
Molecular weight: 34.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
656 |
314 |
342 |
328.0 |
36.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.555 ± 0.504 | 1.22 ± 0.381 |
6.402 ± 1.481 | 3.659 ± 0.727 |
3.963 ± 0.301 | 9.146 ± 0.567 |
2.287 ± 0.663 | 3.354 ± 0.736 |
3.354 ± 0.723 | 7.47 ± 0.322 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.896 ± 0.228 | 4.726 ± 0.242 |
3.659 ± 0.936 | 2.896 ± 0.814 |
9.146 ± 0.058 | 10.366 ± 2.179 |
6.25 ± 1.955 | 6.402 ± 0.647 |
2.439 ± 0.346 | 3.811 ± 0.215 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
