
Pomacea canaliculata (Golden apple snail)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Mollusca; Gastropoda; Caenogastropoda; Architaenioglossa; Ampullarioidea; Ampullariidae; Pomacea
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
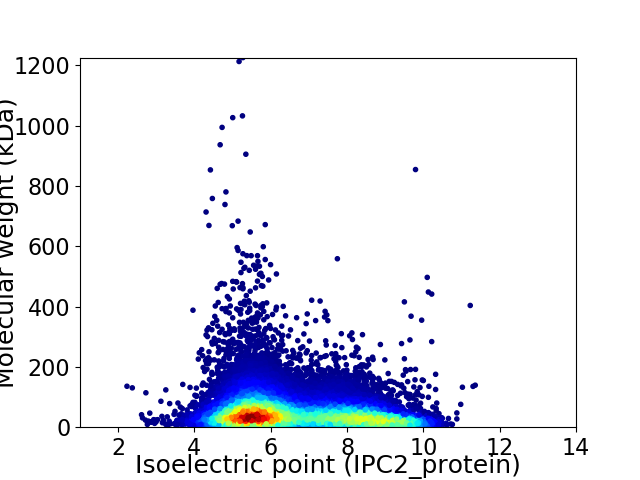
Virtual 2D-PAGE plot for 21377 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2T7PYU5|A0A2T7PYU5_POMCA Uncharacterized protein OS=Pomacea canaliculata OX=400727 GN=C0Q70_01194 PE=4 SV=1
MM1 pKa = 8.19DD2 pKa = 5.07GLRR5 pKa = 11.84QAGHH9 pKa = 5.87NWGSPHH15 pKa = 7.62DD16 pKa = 3.88PTALGEE22 pKa = 4.21CAPNDD27 pKa = 3.95TSGGRR32 pKa = 11.84YY33 pKa = 8.97IMYY36 pKa = 9.51PSLSQGNQPNNFRR49 pKa = 11.84FSPCSIDD56 pKa = 3.63YY57 pKa = 10.3ISPVLRR63 pKa = 11.84DD64 pKa = 3.42ASQEE68 pKa = 4.04CFIEE72 pKa = 4.33SAQDD76 pKa = 3.58EE77 pKa = 4.65IHH79 pKa = 6.79CGNGVVDD86 pKa = 4.53IRR88 pKa = 11.84EE89 pKa = 4.1EE90 pKa = 4.2CDD92 pKa = 3.07AGPDD96 pKa = 3.66GLEE99 pKa = 4.64GKK101 pKa = 9.55DD102 pKa = 3.46KK103 pKa = 10.71CCSSICLLRR112 pKa = 11.84EE113 pKa = 3.75NARR116 pKa = 11.84CRR118 pKa = 11.84QFCLLLLKK126 pKa = 10.68GVLDD130 pKa = 6.09DD131 pKa = 6.94DD132 pKa = 6.56DD133 pKa = 7.6DD134 pKa = 7.66DD135 pKa = 7.66DD136 pKa = 7.69DD137 pKa = 7.65DD138 pKa = 7.65DD139 pKa = 7.65DD140 pKa = 7.65DD141 pKa = 7.65DD142 pKa = 7.65DD143 pKa = 7.65DD144 pKa = 7.68DD145 pKa = 7.67DD146 pKa = 7.63DD147 pKa = 7.61DD148 pKa = 7.17DD149 pKa = 5.09DD150 pKa = 4.23
MM1 pKa = 8.19DD2 pKa = 5.07GLRR5 pKa = 11.84QAGHH9 pKa = 5.87NWGSPHH15 pKa = 7.62DD16 pKa = 3.88PTALGEE22 pKa = 4.21CAPNDD27 pKa = 3.95TSGGRR32 pKa = 11.84YY33 pKa = 8.97IMYY36 pKa = 9.51PSLSQGNQPNNFRR49 pKa = 11.84FSPCSIDD56 pKa = 3.63YY57 pKa = 10.3ISPVLRR63 pKa = 11.84DD64 pKa = 3.42ASQEE68 pKa = 4.04CFIEE72 pKa = 4.33SAQDD76 pKa = 3.58EE77 pKa = 4.65IHH79 pKa = 6.79CGNGVVDD86 pKa = 4.53IRR88 pKa = 11.84EE89 pKa = 4.1EE90 pKa = 4.2CDD92 pKa = 3.07AGPDD96 pKa = 3.66GLEE99 pKa = 4.64GKK101 pKa = 9.55DD102 pKa = 3.46KK103 pKa = 10.71CCSSICLLRR112 pKa = 11.84EE113 pKa = 3.75NARR116 pKa = 11.84CRR118 pKa = 11.84QFCLLLLKK126 pKa = 10.68GVLDD130 pKa = 6.09DD131 pKa = 6.94DD132 pKa = 6.56DD133 pKa = 7.6DD134 pKa = 7.66DD135 pKa = 7.66DD136 pKa = 7.69DD137 pKa = 7.65DD138 pKa = 7.65DD139 pKa = 7.65DD140 pKa = 7.65DD141 pKa = 7.65DD142 pKa = 7.65DD143 pKa = 7.65DD144 pKa = 7.68DD145 pKa = 7.67DD146 pKa = 7.63DD147 pKa = 7.61DD148 pKa = 7.17DD149 pKa = 5.09DD150 pKa = 4.23
Molecular weight: 16.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T7PKI8|A0A2T7PKI8_POMCA t-SNARE coiled-coil homology domain-containing protein OS=Pomacea canaliculata OX=400727 GN=C0Q70_05182 PE=3 SV=1
MM1 pKa = 7.29PRR3 pKa = 11.84NVKK6 pKa = 10.31DD7 pKa = 3.46HH8 pKa = 6.54GCPIIVSSSFTANTGTLTLQAFLRR32 pKa = 11.84SIKK35 pKa = 10.45NPEE38 pKa = 3.64SAADD42 pKa = 3.5INQGTNSSDD51 pKa = 2.74IYY53 pKa = 10.94SVVRR57 pKa = 11.84PRR59 pKa = 11.84RR60 pKa = 11.84GAPARR65 pKa = 11.84VFSQTTTSNKK75 pKa = 6.87QTTAALRR82 pKa = 11.84RR83 pKa = 11.84ILTTTTKK90 pKa = 7.74TTHH93 pKa = 5.87RR94 pKa = 11.84TNATLKK100 pKa = 9.83QATATVYY107 pKa = 8.62RR108 pKa = 11.84TTTAKK113 pKa = 10.28PSRR116 pKa = 11.84LGVTTRR122 pKa = 11.84PTTKK126 pKa = 7.43TTRR129 pKa = 11.84RR130 pKa = 11.84GIATSQTTKK139 pKa = 7.86TTWRR143 pKa = 11.84GITTRR148 pKa = 11.84PTTKK152 pKa = 8.13TTRR155 pKa = 11.84LGVTTRR161 pKa = 11.84PTTKK165 pKa = 7.61TTRR168 pKa = 11.84IGVTTRR174 pKa = 11.84PTTRR178 pKa = 11.84TTRR181 pKa = 11.84IGVTTRR187 pKa = 11.84PATRR191 pKa = 11.84TTRR194 pKa = 11.84LGVTTKK200 pKa = 8.1TTTKK204 pKa = 6.06TTRR207 pKa = 11.84IGVTTRR213 pKa = 11.84PTTKK217 pKa = 10.66ANATLKK223 pKa = 8.91PTTKK227 pKa = 9.07TVQRR231 pKa = 11.84ITTAKK236 pKa = 5.46TTRR239 pKa = 11.84LGVTTRR245 pKa = 11.84PTTRR249 pKa = 11.84TTRR252 pKa = 11.84LGVTTRR258 pKa = 11.84PTTRR262 pKa = 11.84TTRR265 pKa = 11.84LGVTTKK271 pKa = 8.1TTTKK275 pKa = 6.06TTRR278 pKa = 11.84IGVTTKK284 pKa = 9.14PTTKK288 pKa = 7.49TTRR291 pKa = 11.84IGVTTRR297 pKa = 11.84PTTRR301 pKa = 11.84TTRR304 pKa = 11.84LGVTTRR310 pKa = 11.84PTTRR314 pKa = 11.84TTRR317 pKa = 11.84LGITTRR323 pKa = 11.84PTTKK327 pKa = 10.67ANATLKK333 pKa = 8.91PTTKK337 pKa = 9.07TVQRR341 pKa = 11.84ITTAKK346 pKa = 5.46TTRR349 pKa = 11.84LGVTTRR355 pKa = 11.84PTTKK359 pKa = 8.15TTRR362 pKa = 11.84LGITTRR368 pKa = 11.84PTTRR372 pKa = 11.84TTRR375 pKa = 11.84LGITTRR381 pKa = 11.84PTTRR385 pKa = 11.84TTRR388 pKa = 11.84LGITTRR394 pKa = 11.84PTTRR398 pKa = 11.84TTRR401 pKa = 11.84LGITTRR407 pKa = 11.84PTTRR411 pKa = 11.84TTRR414 pKa = 11.84LGITTRR420 pKa = 11.84PTTRR424 pKa = 11.84TTRR427 pKa = 11.84LGVTTRR433 pKa = 11.84PTTRR437 pKa = 11.84TTRR440 pKa = 11.84LGVTTKK446 pKa = 8.1TTTKK450 pKa = 6.06TTRR453 pKa = 11.84IGVTTRR459 pKa = 11.84PTTRR463 pKa = 11.84TTRR466 pKa = 11.84LGVTTRR472 pKa = 11.84PTTRR476 pKa = 11.84TTRR479 pKa = 11.84LGITTRR485 pKa = 11.84QTTKK489 pKa = 11.09ANATLKK495 pKa = 8.91PTTKK499 pKa = 9.07TVQRR503 pKa = 11.84ITTAKK508 pKa = 5.46TTRR511 pKa = 11.84LGVTTRR517 pKa = 11.84PTIRR521 pKa = 11.84TTRR524 pKa = 11.84LGVTTRR530 pKa = 11.84PTTRR534 pKa = 11.84TTRR537 pKa = 11.84LGITTRR543 pKa = 11.84PTTRR547 pKa = 11.84TTRR550 pKa = 11.84IRR552 pKa = 11.84VTTRR556 pKa = 11.84PTTRR560 pKa = 11.84TTRR563 pKa = 11.84LGITTRR569 pKa = 11.84PTTRR573 pKa = 11.84TTRR576 pKa = 11.84LGITTRR582 pKa = 11.84PTTRR586 pKa = 11.84TTRR589 pKa = 11.84LGITTRR595 pKa = 11.84PTTKK599 pKa = 8.17TTRR602 pKa = 11.84LGITTRR608 pKa = 11.84PTTRR612 pKa = 11.84TTRR615 pKa = 11.84LGITTRR621 pKa = 11.84PTTRR625 pKa = 11.84TTRR628 pKa = 11.84LGITTRR634 pKa = 11.84PTTRR638 pKa = 11.84TTRR641 pKa = 11.84LGVTTRR647 pKa = 11.84PTTRR651 pKa = 11.84TTRR654 pKa = 11.84LGITTKK660 pKa = 9.26PTTRR664 pKa = 11.84TTRR667 pKa = 11.84LGVTTRR673 pKa = 11.84PTTRR677 pKa = 11.84TTRR680 pKa = 11.84LGVTTRR686 pKa = 11.84PTIRR690 pKa = 11.84TTRR693 pKa = 11.84LGITTRR699 pKa = 11.84PTTKK703 pKa = 8.17TTRR706 pKa = 11.84LGITTRR712 pKa = 11.84PTTKK716 pKa = 8.17TTRR719 pKa = 11.84LGITTRR725 pKa = 11.84PTTKK729 pKa = 10.22TNATLKK735 pKa = 8.77PTTKK739 pKa = 9.22TVQSITTAKK748 pKa = 6.0TTRR751 pKa = 11.84LGVTTRR757 pKa = 11.84PTTRR761 pKa = 11.84TTRR764 pKa = 11.84LGVTTRR770 pKa = 11.84PTTRR774 pKa = 11.84TTRR777 pKa = 11.84LGVTTRR783 pKa = 11.84PTTRR787 pKa = 11.84TTRR790 pKa = 11.84LGVTTRR796 pKa = 11.84PTTRR800 pKa = 11.84TTRR803 pKa = 11.84LGVITRR809 pKa = 11.84PTTRR813 pKa = 11.84TTRR816 pKa = 11.84LGVTTRR822 pKa = 11.84PSTRR826 pKa = 11.84TTRR829 pKa = 11.84LGVTTRR835 pKa = 11.84PTTRR839 pKa = 11.84TTRR842 pKa = 11.84LGITTRR848 pKa = 11.84PTTKK852 pKa = 10.22TNATLKK858 pKa = 8.77PTTKK862 pKa = 9.22TVQSITTAKK871 pKa = 6.0TTRR874 pKa = 11.84LGVTTRR880 pKa = 11.84PTTRR884 pKa = 11.84TTRR887 pKa = 11.84IGVTTRR893 pKa = 11.84AITKK897 pKa = 6.41TTLPRR902 pKa = 11.84VSARR906 pKa = 11.84PTAKK910 pKa = 10.03TNTTLKK916 pKa = 8.99PTTKK920 pKa = 9.06TVQSITTAKK929 pKa = 6.0TTRR932 pKa = 11.84LGVTTRR938 pKa = 11.84PTTRR942 pKa = 11.84TTRR945 pKa = 11.84IGVTTRR951 pKa = 11.84AITKK955 pKa = 6.41TTLPRR960 pKa = 11.84VSARR964 pKa = 11.84PTAKK968 pKa = 10.07TNTTLKK974 pKa = 9.56STTKK978 pKa = 8.93TVQSITTAKK987 pKa = 5.58TTRR990 pKa = 11.84PRR992 pKa = 11.84VSARR996 pKa = 11.84TTTKK1000 pKa = 7.6TTKK1003 pKa = 9.62LTTQKK1008 pKa = 7.25TTRR1011 pKa = 11.84TRR1013 pKa = 11.84VTTRR1017 pKa = 11.84PTTKK1021 pKa = 8.92STRR1024 pKa = 11.84PRR1026 pKa = 11.84VTSRR1030 pKa = 11.84PTTNTTRR1037 pKa = 11.84PTTQKK1042 pKa = 6.6TTRR1045 pKa = 11.84TRR1047 pKa = 11.84VTTRR1051 pKa = 11.84RR1052 pKa = 11.84TTKK1055 pKa = 6.09TTRR1058 pKa = 11.84PRR1060 pKa = 11.84VTSRR1064 pKa = 11.84RR1065 pKa = 11.84TTKK1068 pKa = 7.26TTRR1071 pKa = 11.84PTTQKK1076 pKa = 8.94STRR1079 pKa = 11.84VTIRR1083 pKa = 11.84RR1084 pKa = 11.84TSKK1087 pKa = 6.56TTRR1090 pKa = 11.84PTTQKK1095 pKa = 8.86STRR1098 pKa = 11.84VTTRR1102 pKa = 11.84RR1103 pKa = 11.84TTKK1106 pKa = 6.72TTRR1109 pKa = 11.84PTTQKK1114 pKa = 6.21TTRR1117 pKa = 11.84VTTRR1121 pKa = 11.84RR1122 pKa = 11.84TTKK1125 pKa = 6.09TTRR1128 pKa = 11.84PRR1130 pKa = 11.84VTSRR1134 pKa = 11.84RR1135 pKa = 11.84TTKK1138 pKa = 7.26TTRR1141 pKa = 11.84PTTQKK1146 pKa = 6.6TTRR1149 pKa = 11.84TRR1151 pKa = 11.84VTARR1155 pKa = 11.84PTTKK1159 pKa = 8.05TTRR1162 pKa = 11.84PTTHH1166 pKa = 6.6KK1167 pKa = 10.42AARR1170 pKa = 11.84TRR1172 pKa = 11.84GTTRR1176 pKa = 11.84PTTKK1180 pKa = 7.47TTRR1183 pKa = 11.84RR1184 pKa = 11.84GVNARR1189 pKa = 11.84ITTPKK1194 pKa = 10.38VKK1196 pKa = 9.95TPAQTAAPLLSEE1208 pKa = 4.81RR1209 pKa = 11.84STEE1212 pKa = 4.28GKK1214 pKa = 9.5SLRR1217 pKa = 11.84SDD1219 pKa = 2.97RR1220 pKa = 11.84GGEE1223 pKa = 3.72LRR1225 pKa = 11.84AVVGVTVGVLGLVIIAALIYY1245 pKa = 9.16FLRR1248 pKa = 11.84KK1249 pKa = 9.28RR1250 pKa = 11.84RR1251 pKa = 11.84IDD1253 pKa = 3.54ANVGDD1258 pKa = 4.95IYY1260 pKa = 11.54NVDD1263 pKa = 3.51LL1264 pKa = 4.89
MM1 pKa = 7.29PRR3 pKa = 11.84NVKK6 pKa = 10.31DD7 pKa = 3.46HH8 pKa = 6.54GCPIIVSSSFTANTGTLTLQAFLRR32 pKa = 11.84SIKK35 pKa = 10.45NPEE38 pKa = 3.64SAADD42 pKa = 3.5INQGTNSSDD51 pKa = 2.74IYY53 pKa = 10.94SVVRR57 pKa = 11.84PRR59 pKa = 11.84RR60 pKa = 11.84GAPARR65 pKa = 11.84VFSQTTTSNKK75 pKa = 6.87QTTAALRR82 pKa = 11.84RR83 pKa = 11.84ILTTTTKK90 pKa = 7.74TTHH93 pKa = 5.87RR94 pKa = 11.84TNATLKK100 pKa = 9.83QATATVYY107 pKa = 8.62RR108 pKa = 11.84TTTAKK113 pKa = 10.28PSRR116 pKa = 11.84LGVTTRR122 pKa = 11.84PTTKK126 pKa = 7.43TTRR129 pKa = 11.84RR130 pKa = 11.84GIATSQTTKK139 pKa = 7.86TTWRR143 pKa = 11.84GITTRR148 pKa = 11.84PTTKK152 pKa = 8.13TTRR155 pKa = 11.84LGVTTRR161 pKa = 11.84PTTKK165 pKa = 7.61TTRR168 pKa = 11.84IGVTTRR174 pKa = 11.84PTTRR178 pKa = 11.84TTRR181 pKa = 11.84IGVTTRR187 pKa = 11.84PATRR191 pKa = 11.84TTRR194 pKa = 11.84LGVTTKK200 pKa = 8.1TTTKK204 pKa = 6.06TTRR207 pKa = 11.84IGVTTRR213 pKa = 11.84PTTKK217 pKa = 10.66ANATLKK223 pKa = 8.91PTTKK227 pKa = 9.07TVQRR231 pKa = 11.84ITTAKK236 pKa = 5.46TTRR239 pKa = 11.84LGVTTRR245 pKa = 11.84PTTRR249 pKa = 11.84TTRR252 pKa = 11.84LGVTTRR258 pKa = 11.84PTTRR262 pKa = 11.84TTRR265 pKa = 11.84LGVTTKK271 pKa = 8.1TTTKK275 pKa = 6.06TTRR278 pKa = 11.84IGVTTKK284 pKa = 9.14PTTKK288 pKa = 7.49TTRR291 pKa = 11.84IGVTTRR297 pKa = 11.84PTTRR301 pKa = 11.84TTRR304 pKa = 11.84LGVTTRR310 pKa = 11.84PTTRR314 pKa = 11.84TTRR317 pKa = 11.84LGITTRR323 pKa = 11.84PTTKK327 pKa = 10.67ANATLKK333 pKa = 8.91PTTKK337 pKa = 9.07TVQRR341 pKa = 11.84ITTAKK346 pKa = 5.46TTRR349 pKa = 11.84LGVTTRR355 pKa = 11.84PTTKK359 pKa = 8.15TTRR362 pKa = 11.84LGITTRR368 pKa = 11.84PTTRR372 pKa = 11.84TTRR375 pKa = 11.84LGITTRR381 pKa = 11.84PTTRR385 pKa = 11.84TTRR388 pKa = 11.84LGITTRR394 pKa = 11.84PTTRR398 pKa = 11.84TTRR401 pKa = 11.84LGITTRR407 pKa = 11.84PTTRR411 pKa = 11.84TTRR414 pKa = 11.84LGITTRR420 pKa = 11.84PTTRR424 pKa = 11.84TTRR427 pKa = 11.84LGVTTRR433 pKa = 11.84PTTRR437 pKa = 11.84TTRR440 pKa = 11.84LGVTTKK446 pKa = 8.1TTTKK450 pKa = 6.06TTRR453 pKa = 11.84IGVTTRR459 pKa = 11.84PTTRR463 pKa = 11.84TTRR466 pKa = 11.84LGVTTRR472 pKa = 11.84PTTRR476 pKa = 11.84TTRR479 pKa = 11.84LGITTRR485 pKa = 11.84QTTKK489 pKa = 11.09ANATLKK495 pKa = 8.91PTTKK499 pKa = 9.07TVQRR503 pKa = 11.84ITTAKK508 pKa = 5.46TTRR511 pKa = 11.84LGVTTRR517 pKa = 11.84PTIRR521 pKa = 11.84TTRR524 pKa = 11.84LGVTTRR530 pKa = 11.84PTTRR534 pKa = 11.84TTRR537 pKa = 11.84LGITTRR543 pKa = 11.84PTTRR547 pKa = 11.84TTRR550 pKa = 11.84IRR552 pKa = 11.84VTTRR556 pKa = 11.84PTTRR560 pKa = 11.84TTRR563 pKa = 11.84LGITTRR569 pKa = 11.84PTTRR573 pKa = 11.84TTRR576 pKa = 11.84LGITTRR582 pKa = 11.84PTTRR586 pKa = 11.84TTRR589 pKa = 11.84LGITTRR595 pKa = 11.84PTTKK599 pKa = 8.17TTRR602 pKa = 11.84LGITTRR608 pKa = 11.84PTTRR612 pKa = 11.84TTRR615 pKa = 11.84LGITTRR621 pKa = 11.84PTTRR625 pKa = 11.84TTRR628 pKa = 11.84LGITTRR634 pKa = 11.84PTTRR638 pKa = 11.84TTRR641 pKa = 11.84LGVTTRR647 pKa = 11.84PTTRR651 pKa = 11.84TTRR654 pKa = 11.84LGITTKK660 pKa = 9.26PTTRR664 pKa = 11.84TTRR667 pKa = 11.84LGVTTRR673 pKa = 11.84PTTRR677 pKa = 11.84TTRR680 pKa = 11.84LGVTTRR686 pKa = 11.84PTIRR690 pKa = 11.84TTRR693 pKa = 11.84LGITTRR699 pKa = 11.84PTTKK703 pKa = 8.17TTRR706 pKa = 11.84LGITTRR712 pKa = 11.84PTTKK716 pKa = 8.17TTRR719 pKa = 11.84LGITTRR725 pKa = 11.84PTTKK729 pKa = 10.22TNATLKK735 pKa = 8.77PTTKK739 pKa = 9.22TVQSITTAKK748 pKa = 6.0TTRR751 pKa = 11.84LGVTTRR757 pKa = 11.84PTTRR761 pKa = 11.84TTRR764 pKa = 11.84LGVTTRR770 pKa = 11.84PTTRR774 pKa = 11.84TTRR777 pKa = 11.84LGVTTRR783 pKa = 11.84PTTRR787 pKa = 11.84TTRR790 pKa = 11.84LGVTTRR796 pKa = 11.84PTTRR800 pKa = 11.84TTRR803 pKa = 11.84LGVITRR809 pKa = 11.84PTTRR813 pKa = 11.84TTRR816 pKa = 11.84LGVTTRR822 pKa = 11.84PSTRR826 pKa = 11.84TTRR829 pKa = 11.84LGVTTRR835 pKa = 11.84PTTRR839 pKa = 11.84TTRR842 pKa = 11.84LGITTRR848 pKa = 11.84PTTKK852 pKa = 10.22TNATLKK858 pKa = 8.77PTTKK862 pKa = 9.22TVQSITTAKK871 pKa = 6.0TTRR874 pKa = 11.84LGVTTRR880 pKa = 11.84PTTRR884 pKa = 11.84TTRR887 pKa = 11.84IGVTTRR893 pKa = 11.84AITKK897 pKa = 6.41TTLPRR902 pKa = 11.84VSARR906 pKa = 11.84PTAKK910 pKa = 10.03TNTTLKK916 pKa = 8.99PTTKK920 pKa = 9.06TVQSITTAKK929 pKa = 6.0TTRR932 pKa = 11.84LGVTTRR938 pKa = 11.84PTTRR942 pKa = 11.84TTRR945 pKa = 11.84IGVTTRR951 pKa = 11.84AITKK955 pKa = 6.41TTLPRR960 pKa = 11.84VSARR964 pKa = 11.84PTAKK968 pKa = 10.07TNTTLKK974 pKa = 9.56STTKK978 pKa = 8.93TVQSITTAKK987 pKa = 5.58TTRR990 pKa = 11.84PRR992 pKa = 11.84VSARR996 pKa = 11.84TTTKK1000 pKa = 7.6TTKK1003 pKa = 9.62LTTQKK1008 pKa = 7.25TTRR1011 pKa = 11.84TRR1013 pKa = 11.84VTTRR1017 pKa = 11.84PTTKK1021 pKa = 8.92STRR1024 pKa = 11.84PRR1026 pKa = 11.84VTSRR1030 pKa = 11.84PTTNTTRR1037 pKa = 11.84PTTQKK1042 pKa = 6.6TTRR1045 pKa = 11.84TRR1047 pKa = 11.84VTTRR1051 pKa = 11.84RR1052 pKa = 11.84TTKK1055 pKa = 6.09TTRR1058 pKa = 11.84PRR1060 pKa = 11.84VTSRR1064 pKa = 11.84RR1065 pKa = 11.84TTKK1068 pKa = 7.26TTRR1071 pKa = 11.84PTTQKK1076 pKa = 8.94STRR1079 pKa = 11.84VTIRR1083 pKa = 11.84RR1084 pKa = 11.84TSKK1087 pKa = 6.56TTRR1090 pKa = 11.84PTTQKK1095 pKa = 8.86STRR1098 pKa = 11.84VTTRR1102 pKa = 11.84RR1103 pKa = 11.84TTKK1106 pKa = 6.72TTRR1109 pKa = 11.84PTTQKK1114 pKa = 6.21TTRR1117 pKa = 11.84VTTRR1121 pKa = 11.84RR1122 pKa = 11.84TTKK1125 pKa = 6.09TTRR1128 pKa = 11.84PRR1130 pKa = 11.84VTSRR1134 pKa = 11.84RR1135 pKa = 11.84TTKK1138 pKa = 7.26TTRR1141 pKa = 11.84PTTQKK1146 pKa = 6.6TTRR1149 pKa = 11.84TRR1151 pKa = 11.84VTARR1155 pKa = 11.84PTTKK1159 pKa = 8.05TTRR1162 pKa = 11.84PTTHH1166 pKa = 6.6KK1167 pKa = 10.42AARR1170 pKa = 11.84TRR1172 pKa = 11.84GTTRR1176 pKa = 11.84PTTKK1180 pKa = 7.47TTRR1183 pKa = 11.84RR1184 pKa = 11.84GVNARR1189 pKa = 11.84ITTPKK1194 pKa = 10.38VKK1196 pKa = 9.95TPAQTAAPLLSEE1208 pKa = 4.81RR1209 pKa = 11.84STEE1212 pKa = 4.28GKK1214 pKa = 9.5SLRR1217 pKa = 11.84SDD1219 pKa = 2.97RR1220 pKa = 11.84GGEE1223 pKa = 3.72LRR1225 pKa = 11.84AVVGVTVGVLGLVIIAALIYY1245 pKa = 9.16FLRR1248 pKa = 11.84KK1249 pKa = 9.28RR1250 pKa = 11.84RR1251 pKa = 11.84IDD1253 pKa = 3.54ANVGDD1258 pKa = 4.95IYY1260 pKa = 11.54NVDD1263 pKa = 3.51LL1264 pKa = 4.89
Molecular weight: 139.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10621572 |
52 |
10929 |
496.9 |
55.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.776 ± 0.017 | 2.322 ± 0.019 |
5.598 ± 0.017 | 6.27 ± 0.024 |
3.624 ± 0.012 | 6.188 ± 0.032 |
2.698 ± 0.011 | 4.476 ± 0.013 |
5.375 ± 0.022 | 9.064 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.337 ± 0.007 | 4.025 ± 0.013 |
5.235 ± 0.025 | 4.643 ± 0.018 |
5.718 ± 0.017 | 8.696 ± 0.028 |
6.227 ± 0.02 | 6.821 ± 0.018 |
1.165 ± 0.008 | 2.741 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
