
Capybara microvirus Cap1_SP_108
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
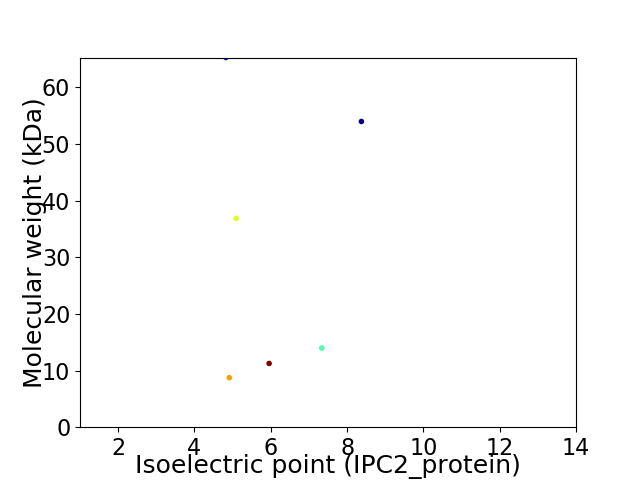
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A4P8W4E9|A0A4P8W4E9_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_108 OX=2584806 PE=4 SV=1
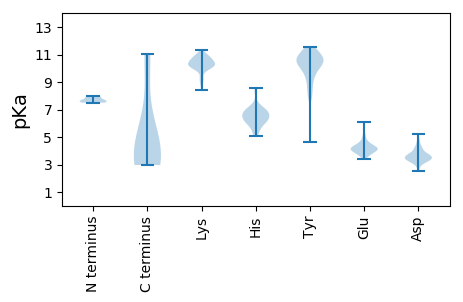
MM1 pKa = 7.66NIYY4 pKa = 10.35SIYY7 pKa = 11.29DD8 pKa = 3.2MVARR12 pKa = 11.84EE13 pKa = 3.97FSSPFLAKK21 pKa = 10.22NDD23 pKa = 3.58GVAMRR28 pKa = 11.84MFGNTCRR35 pKa = 11.84SEE37 pKa = 5.12RR38 pKa = 11.84IVNPNEE44 pKa = 3.69LEE46 pKa = 4.17LYY48 pKa = 10.62CIGKK52 pKa = 9.32FLGEE56 pKa = 4.2NLDD59 pKa = 4.19GCIEE63 pKa = 4.04PCKK66 pKa = 10.39PMIIRR71 pKa = 11.84VSEE74 pKa = 4.09SDD76 pKa = 3.33VV77 pKa = 3.24
MM1 pKa = 7.66NIYY4 pKa = 10.35SIYY7 pKa = 11.29DD8 pKa = 3.2MVARR12 pKa = 11.84EE13 pKa = 3.97FSSPFLAKK21 pKa = 10.22NDD23 pKa = 3.58GVAMRR28 pKa = 11.84MFGNTCRR35 pKa = 11.84SEE37 pKa = 5.12RR38 pKa = 11.84IVNPNEE44 pKa = 3.69LEE46 pKa = 4.17LYY48 pKa = 10.62CIGKK52 pKa = 9.32FLGEE56 pKa = 4.2NLDD59 pKa = 4.19GCIEE63 pKa = 4.04PCKK66 pKa = 10.39PMIIRR71 pKa = 11.84VSEE74 pKa = 4.09SDD76 pKa = 3.33VV77 pKa = 3.24
Molecular weight: 8.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W3Y6|A0A4P8W3Y6_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_108 OX=2584806 PE=3 SV=1
MM1 pKa = 8.01PCGQCVNCRR10 pKa = 11.84KK11 pKa = 10.22RR12 pKa = 11.84IRR14 pKa = 11.84SQWTGRR20 pKa = 11.84MMMEE24 pKa = 4.18NLTHH28 pKa = 6.72KK29 pKa = 10.62ASVFITLSYY38 pKa = 11.4DD39 pKa = 3.64DD40 pKa = 3.74VHH42 pKa = 8.58LPIIKK47 pKa = 9.84INEE50 pKa = 4.05RR51 pKa = 11.84SFTSYY56 pKa = 11.09DD57 pKa = 3.29SEE59 pKa = 4.06LTYY62 pKa = 10.55RR63 pKa = 11.84DD64 pKa = 4.16CCCLDD69 pKa = 3.27AHH71 pKa = 6.82NILHH75 pKa = 6.75PVRR78 pKa = 11.84LPIFSSTDD86 pKa = 3.02TSDD89 pKa = 5.22GIISDD94 pKa = 4.64FASCRR99 pKa = 11.84PNTFLTDD106 pKa = 3.59YY107 pKa = 11.04SNSPCVDD114 pKa = 5.04EE115 pKa = 5.42IPVLAPLQPRR125 pKa = 11.84DD126 pKa = 3.4TQLFLKK132 pKa = 10.36RR133 pKa = 11.84LRR135 pKa = 11.84INLKK139 pKa = 10.03RR140 pKa = 11.84QFGLSDD146 pKa = 3.16KK147 pKa = 10.79LRR149 pKa = 11.84YY150 pKa = 8.59FLCGEE155 pKa = 4.09YY156 pKa = 10.76GEE158 pKa = 4.6EE159 pKa = 3.75RR160 pKa = 11.84GRR162 pKa = 11.84PHH164 pKa = 7.17YY165 pKa = 10.94HH166 pKa = 6.41MILFGLDD173 pKa = 3.49VSPSSYY179 pKa = 10.47RR180 pKa = 11.84CRR182 pKa = 11.84DD183 pKa = 3.27SNSYY187 pKa = 10.32SKK189 pKa = 10.91AYY191 pKa = 10.52ALDD194 pKa = 3.57SVFRR198 pKa = 11.84CIQDD202 pKa = 2.65SWSLGRR208 pKa = 11.84IEE210 pKa = 4.98CEE212 pKa = 3.55PVTEE216 pKa = 4.39SNLRR220 pKa = 11.84YY221 pKa = 8.13VAKK224 pKa = 9.28YY225 pKa = 4.67TTKK228 pKa = 10.55TSMDD232 pKa = 3.81SAPSIEE238 pKa = 3.6WDD240 pKa = 3.67YY241 pKa = 11.55YY242 pKa = 10.32IRR244 pKa = 11.84QCLNYY249 pKa = 9.94LKK251 pKa = 10.3HH252 pKa = 6.03NCPMKK257 pKa = 10.62CDD259 pKa = 3.73SFCFDD264 pKa = 2.86SAKK267 pKa = 10.48RR268 pKa = 11.84FLRR271 pKa = 11.84SLRR274 pKa = 11.84DD275 pKa = 3.36FDD277 pKa = 4.15IYY279 pKa = 10.62PYY281 pKa = 9.8GQKK284 pKa = 10.12PFHH287 pKa = 6.27RR288 pKa = 11.84QSLGLGYY295 pKa = 10.72DD296 pKa = 3.73FFYY299 pKa = 11.45KK300 pKa = 10.74NIDD303 pKa = 3.71SFAHH307 pKa = 6.02KK308 pKa = 9.95PYY310 pKa = 9.25FTSASSKK317 pKa = 9.93IRR319 pKa = 11.84YY320 pKa = 7.88SFPKK324 pKa = 10.19CFYY327 pKa = 10.73DD328 pKa = 3.49RR329 pKa = 11.84YY330 pKa = 10.71KK331 pKa = 10.16RR332 pKa = 11.84TTFRR336 pKa = 11.84LYY338 pKa = 10.8QNSLLQKK345 pKa = 10.08YY346 pKa = 9.78DD347 pKa = 3.33FEE349 pKa = 5.98SYY351 pKa = 7.82VHH353 pKa = 6.81CCNNMWSSLYY363 pKa = 10.0EE364 pKa = 4.31SKK366 pKa = 10.92GLNYY370 pKa = 10.34RR371 pKa = 11.84PIFPSAKK378 pKa = 9.92IKK380 pKa = 10.87DD381 pKa = 4.0LFLKK385 pKa = 10.51SRR387 pKa = 11.84IYY389 pKa = 11.02GSFSNDD395 pKa = 3.11YY396 pKa = 9.56FPSFRR401 pKa = 11.84SPVPSYY407 pKa = 10.96KK408 pKa = 10.08KK409 pKa = 9.65WFEE412 pKa = 4.28SYY414 pKa = 10.56SKK416 pKa = 10.91HH417 pKa = 5.71NLWHH421 pKa = 6.44KK422 pKa = 10.25MPVLAHH428 pKa = 5.99SSVFVMFKK436 pKa = 10.51PPVSPPSYY444 pKa = 11.35NEE446 pKa = 3.97FCSLTYY452 pKa = 10.71KK453 pKa = 10.11HH454 pKa = 6.6LKK456 pKa = 9.15RR457 pKa = 4.93
MM1 pKa = 8.01PCGQCVNCRR10 pKa = 11.84KK11 pKa = 10.22RR12 pKa = 11.84IRR14 pKa = 11.84SQWTGRR20 pKa = 11.84MMMEE24 pKa = 4.18NLTHH28 pKa = 6.72KK29 pKa = 10.62ASVFITLSYY38 pKa = 11.4DD39 pKa = 3.64DD40 pKa = 3.74VHH42 pKa = 8.58LPIIKK47 pKa = 9.84INEE50 pKa = 4.05RR51 pKa = 11.84SFTSYY56 pKa = 11.09DD57 pKa = 3.29SEE59 pKa = 4.06LTYY62 pKa = 10.55RR63 pKa = 11.84DD64 pKa = 4.16CCCLDD69 pKa = 3.27AHH71 pKa = 6.82NILHH75 pKa = 6.75PVRR78 pKa = 11.84LPIFSSTDD86 pKa = 3.02TSDD89 pKa = 5.22GIISDD94 pKa = 4.64FASCRR99 pKa = 11.84PNTFLTDD106 pKa = 3.59YY107 pKa = 11.04SNSPCVDD114 pKa = 5.04EE115 pKa = 5.42IPVLAPLQPRR125 pKa = 11.84DD126 pKa = 3.4TQLFLKK132 pKa = 10.36RR133 pKa = 11.84LRR135 pKa = 11.84INLKK139 pKa = 10.03RR140 pKa = 11.84QFGLSDD146 pKa = 3.16KK147 pKa = 10.79LRR149 pKa = 11.84YY150 pKa = 8.59FLCGEE155 pKa = 4.09YY156 pKa = 10.76GEE158 pKa = 4.6EE159 pKa = 3.75RR160 pKa = 11.84GRR162 pKa = 11.84PHH164 pKa = 7.17YY165 pKa = 10.94HH166 pKa = 6.41MILFGLDD173 pKa = 3.49VSPSSYY179 pKa = 10.47RR180 pKa = 11.84CRR182 pKa = 11.84DD183 pKa = 3.27SNSYY187 pKa = 10.32SKK189 pKa = 10.91AYY191 pKa = 10.52ALDD194 pKa = 3.57SVFRR198 pKa = 11.84CIQDD202 pKa = 2.65SWSLGRR208 pKa = 11.84IEE210 pKa = 4.98CEE212 pKa = 3.55PVTEE216 pKa = 4.39SNLRR220 pKa = 11.84YY221 pKa = 8.13VAKK224 pKa = 9.28YY225 pKa = 4.67TTKK228 pKa = 10.55TSMDD232 pKa = 3.81SAPSIEE238 pKa = 3.6WDD240 pKa = 3.67YY241 pKa = 11.55YY242 pKa = 10.32IRR244 pKa = 11.84QCLNYY249 pKa = 9.94LKK251 pKa = 10.3HH252 pKa = 6.03NCPMKK257 pKa = 10.62CDD259 pKa = 3.73SFCFDD264 pKa = 2.86SAKK267 pKa = 10.48RR268 pKa = 11.84FLRR271 pKa = 11.84SLRR274 pKa = 11.84DD275 pKa = 3.36FDD277 pKa = 4.15IYY279 pKa = 10.62PYY281 pKa = 9.8GQKK284 pKa = 10.12PFHH287 pKa = 6.27RR288 pKa = 11.84QSLGLGYY295 pKa = 10.72DD296 pKa = 3.73FFYY299 pKa = 11.45KK300 pKa = 10.74NIDD303 pKa = 3.71SFAHH307 pKa = 6.02KK308 pKa = 9.95PYY310 pKa = 9.25FTSASSKK317 pKa = 9.93IRR319 pKa = 11.84YY320 pKa = 7.88SFPKK324 pKa = 10.19CFYY327 pKa = 10.73DD328 pKa = 3.49RR329 pKa = 11.84YY330 pKa = 10.71KK331 pKa = 10.16RR332 pKa = 11.84TTFRR336 pKa = 11.84LYY338 pKa = 10.8QNSLLQKK345 pKa = 10.08YY346 pKa = 9.78DD347 pKa = 3.33FEE349 pKa = 5.98SYY351 pKa = 7.82VHH353 pKa = 6.81CCNNMWSSLYY363 pKa = 10.0EE364 pKa = 4.31SKK366 pKa = 10.92GLNYY370 pKa = 10.34RR371 pKa = 11.84PIFPSAKK378 pKa = 9.92IKK380 pKa = 10.87DD381 pKa = 4.0LFLKK385 pKa = 10.51SRR387 pKa = 11.84IYY389 pKa = 11.02GSFSNDD395 pKa = 3.11YY396 pKa = 9.56FPSFRR401 pKa = 11.84SPVPSYY407 pKa = 10.96KK408 pKa = 10.08KK409 pKa = 9.65WFEE412 pKa = 4.28SYY414 pKa = 10.56SKK416 pKa = 10.91HH417 pKa = 5.71NLWHH421 pKa = 6.44KK422 pKa = 10.25MPVLAHH428 pKa = 5.99SSVFVMFKK436 pKa = 10.51PPVSPPSYY444 pKa = 11.35NEE446 pKa = 3.97FCSLTYY452 pKa = 10.71KK453 pKa = 10.11HH454 pKa = 6.6LKK456 pKa = 9.15RR457 pKa = 4.93
Molecular weight: 53.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1669 |
77 |
581 |
278.2 |
31.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.752 ± 1.077 | 2.037 ± 0.865 |
6.291 ± 0.314 | 6.471 ± 1.102 |
6.111 ± 0.691 | 6.052 ± 1.202 |
2.397 ± 0.273 | 5.213 ± 0.456 |
5.632 ± 1.149 | 7.549 ± 0.743 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.636 ± 0.445 | 6.111 ± 0.66 |
4.254 ± 1.026 | 2.217 ± 0.367 |
5.872 ± 0.566 | 9.706 ± 1.393 |
3.954 ± 0.93 | 5.992 ± 0.997 |
1.138 ± 0.222 | 4.614 ± 0.958 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
