
Caldiarchaeum subterraneum
Taxonomy: cellular organisms; Archaea; TACK group; Thaumarchaeota; Thaumarchaeota incertae sedis; Candidatus Caldarchaeum
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins
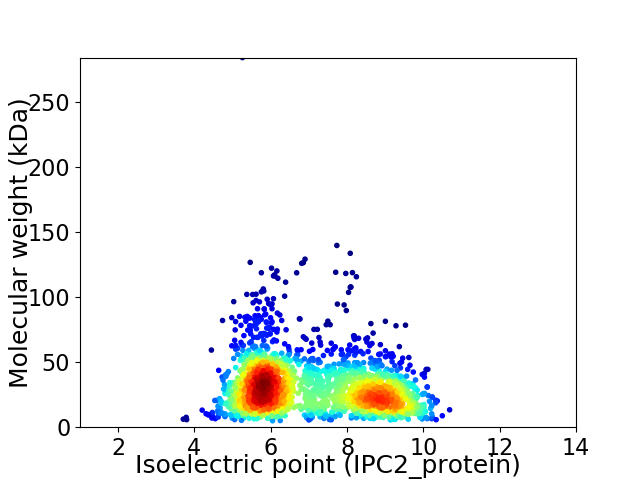
Virtual 2D-PAGE plot for 2154 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
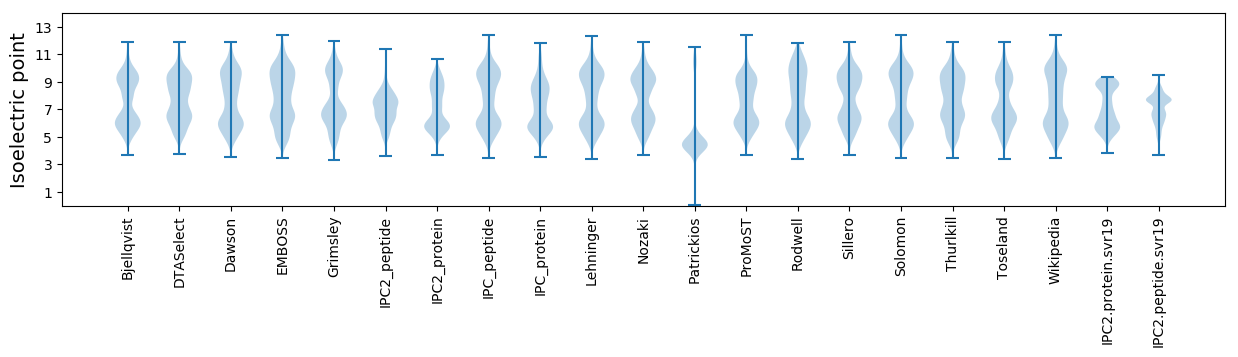
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E6N358|E6N358_CALS0 Thioredoxin reductase OS=Caldiarchaeum subterraneum OX=311458 GN=HGMM_F22F09C14 PE=4 SV=1
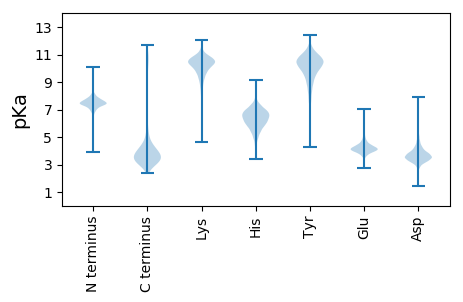
MM1 pKa = 7.31GLEE4 pKa = 4.07PTTPDD9 pKa = 3.69LEE11 pKa = 4.56GRR13 pKa = 11.84CAIHH17 pKa = 5.97SAPRR21 pKa = 11.84ALTVDD26 pKa = 4.08EE27 pKa = 4.52KK28 pKa = 10.98RR29 pKa = 11.84CGVFEE34 pKa = 4.81LSLSVGPSDD43 pKa = 3.7EE44 pKa = 4.68SFFEE48 pKa = 4.1EE49 pKa = 5.82GEE51 pKa = 4.24DD52 pKa = 3.71EE53 pKa = 4.63EE54 pKa = 5.25EE55 pKa = 4.23ADD57 pKa = 4.22SYY59 pKa = 11.45EE60 pKa = 3.95GDD62 pKa = 3.72YY63 pKa = 11.72EE64 pKa = 4.32EE65 pKa = 5.25ACEE68 pKa = 4.55DD69 pKa = 3.45
MM1 pKa = 7.31GLEE4 pKa = 4.07PTTPDD9 pKa = 3.69LEE11 pKa = 4.56GRR13 pKa = 11.84CAIHH17 pKa = 5.97SAPRR21 pKa = 11.84ALTVDD26 pKa = 4.08EE27 pKa = 4.52KK28 pKa = 10.98RR29 pKa = 11.84CGVFEE34 pKa = 4.81LSLSVGPSDD43 pKa = 3.7EE44 pKa = 4.68SFFEE48 pKa = 4.1EE49 pKa = 5.82GEE51 pKa = 4.24DD52 pKa = 3.71EE53 pKa = 4.63EE54 pKa = 5.25EE55 pKa = 4.23ADD57 pKa = 4.22SYY59 pKa = 11.45EE60 pKa = 3.95GDD62 pKa = 3.72YY63 pKa = 11.72EE64 pKa = 4.32EE65 pKa = 5.25ACEE68 pKa = 4.55DD69 pKa = 3.45
Molecular weight: 7.59 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E6N330|E6N330_CALS0 Translation initiation factor eIF-2B OS=Caldiarchaeum subterraneum OX=311458 GN=CSUB_C1164 PE=3 SV=1
MM1 pKa = 8.1SFEE4 pKa = 4.77DD5 pKa = 3.75LNNRR9 pKa = 11.84IISCKK14 pKa = 9.82LCPRR18 pKa = 11.84LVKK21 pKa = 10.58YY22 pKa = 10.55RR23 pKa = 11.84EE24 pKa = 4.05EE25 pKa = 4.17VAKK28 pKa = 10.83NPPKK32 pKa = 10.48RR33 pKa = 11.84FRR35 pKa = 11.84GQTYY39 pKa = 7.14WAKK42 pKa = 9.86PLPGFGDD49 pKa = 3.19VGARR53 pKa = 11.84VLVLGLAPAAHH64 pKa = 6.74GGNRR68 pKa = 11.84TGRR71 pKa = 11.84MFTGDD76 pKa = 3.28SSGDD80 pKa = 3.41TLVRR84 pKa = 11.84ALHH87 pKa = 6.19RR88 pKa = 11.84AGFANMGRR96 pKa = 11.84SISINDD102 pKa = 3.65GLVLKK107 pKa = 10.29DD108 pKa = 3.44VYY110 pKa = 9.39ITAVVRR116 pKa = 11.84CAPPDD121 pKa = 3.59NKK123 pKa = 9.96PAKK126 pKa = 10.18QEE128 pKa = 4.01VEE130 pKa = 3.75NCLPYY135 pKa = 10.64LIEE138 pKa = 4.16EE139 pKa = 4.3LRR141 pKa = 11.84MLEE144 pKa = 4.03NVVVVVALGRR154 pKa = 11.84FAFEE158 pKa = 3.89TFFRR162 pKa = 11.84LMRR165 pKa = 11.84RR166 pKa = 11.84MNAYY170 pKa = 9.58SGRR173 pKa = 11.84IPRR176 pKa = 11.84FRR178 pKa = 11.84HH179 pKa = 4.37GAVYY183 pKa = 10.56RR184 pKa = 11.84LNGSFNGKK192 pKa = 8.76PLPTVIASYY201 pKa = 10.59HH202 pKa = 6.51PSRR205 pKa = 11.84QNTSTGRR212 pKa = 11.84LTQRR216 pKa = 11.84MIDD219 pKa = 3.88SVFHH223 pKa = 6.63RR224 pKa = 11.84ARR226 pKa = 11.84AIAGLL231 pKa = 3.76
MM1 pKa = 8.1SFEE4 pKa = 4.77DD5 pKa = 3.75LNNRR9 pKa = 11.84IISCKK14 pKa = 9.82LCPRR18 pKa = 11.84LVKK21 pKa = 10.58YY22 pKa = 10.55RR23 pKa = 11.84EE24 pKa = 4.05EE25 pKa = 4.17VAKK28 pKa = 10.83NPPKK32 pKa = 10.48RR33 pKa = 11.84FRR35 pKa = 11.84GQTYY39 pKa = 7.14WAKK42 pKa = 9.86PLPGFGDD49 pKa = 3.19VGARR53 pKa = 11.84VLVLGLAPAAHH64 pKa = 6.74GGNRR68 pKa = 11.84TGRR71 pKa = 11.84MFTGDD76 pKa = 3.28SSGDD80 pKa = 3.41TLVRR84 pKa = 11.84ALHH87 pKa = 6.19RR88 pKa = 11.84AGFANMGRR96 pKa = 11.84SISINDD102 pKa = 3.65GLVLKK107 pKa = 10.29DD108 pKa = 3.44VYY110 pKa = 9.39ITAVVRR116 pKa = 11.84CAPPDD121 pKa = 3.59NKK123 pKa = 9.96PAKK126 pKa = 10.18QEE128 pKa = 4.01VEE130 pKa = 3.75NCLPYY135 pKa = 10.64LIEE138 pKa = 4.16EE139 pKa = 4.3LRR141 pKa = 11.84MLEE144 pKa = 4.03NVVVVVALGRR154 pKa = 11.84FAFEE158 pKa = 3.89TFFRR162 pKa = 11.84LMRR165 pKa = 11.84RR166 pKa = 11.84MNAYY170 pKa = 9.58SGRR173 pKa = 11.84IPRR176 pKa = 11.84FRR178 pKa = 11.84HH179 pKa = 4.37GAVYY183 pKa = 10.56RR184 pKa = 11.84LNGSFNGKK192 pKa = 8.76PLPTVIASYY201 pKa = 10.59HH202 pKa = 6.51PSRR205 pKa = 11.84QNTSTGRR212 pKa = 11.84LTQRR216 pKa = 11.84MIDD219 pKa = 3.88SVFHH223 pKa = 6.63RR224 pKa = 11.84ARR226 pKa = 11.84AIAGLL231 pKa = 3.76
Molecular weight: 25.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
631940 |
43 |
2586 |
293.4 |
32.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.465 ± 0.055 | 0.86 ± 0.019 |
4.514 ± 0.034 | 7.067 ± 0.06 |
3.963 ± 0.033 | 7.561 ± 0.047 |
1.806 ± 0.02 | 5.91 ± 0.04 |
5.463 ± 0.046 | 10.576 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.457 ± 0.025 | 2.981 ± 0.029 |
4.631 ± 0.034 | 2.496 ± 0.027 |
6.441 ± 0.041 | 5.879 ± 0.04 |
5.225 ± 0.046 | 9.188 ± 0.047 |
1.106 ± 0.02 | 3.411 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
