
Pseudomonas phage Pf1 (Bacteriophage Pf1)
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Primolicivirus
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins
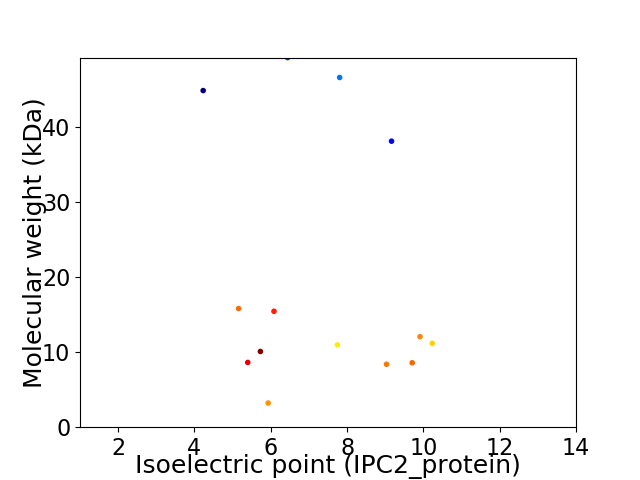
Virtual 2D-PAGE plot for 14 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>sp|P25130|VG430_BPPF1 Uncharacterized protein ORF430 OS=Pseudomonas phage Pf1 OX=2011081 PE=4 SV=1
MM1 pKa = 7.58SIKK4 pKa = 9.83TLISVLRR11 pKa = 11.84VTLLTACLLPSLFFVRR27 pKa = 11.84SAIAGPYY34 pKa = 7.42IWEE37 pKa = 3.97VVMYY41 pKa = 9.58SSSGSSTPAEE51 pKa = 4.3ACEE54 pKa = 4.04KK55 pKa = 10.65ARR57 pKa = 11.84VVADD61 pKa = 5.44RR62 pKa = 11.84SPDD65 pKa = 3.16WNYY68 pKa = 10.64TSATPKK74 pKa = 10.33MNGLDD79 pKa = 3.38NSYY82 pKa = 10.9CSVVYY87 pKa = 9.5VSRR90 pKa = 11.84RR91 pKa = 11.84DD92 pKa = 3.42PSVVNTCDD100 pKa = 4.67DD101 pKa = 3.73CASWKK106 pKa = 10.32LFRR109 pKa = 11.84KK110 pKa = 10.16GDD112 pKa = 3.54QCANADD118 pKa = 3.63DD119 pKa = 4.74TYY121 pKa = 11.35NASTGICEE129 pKa = 4.52PPPKK133 pKa = 9.72EE134 pKa = 4.2CKK136 pKa = 10.06EE137 pKa = 4.24GEE139 pKa = 4.18LFPAKK144 pKa = 10.51GPDD147 pKa = 3.63SPVVTSGGRR156 pKa = 11.84NYY158 pKa = 10.44VGDD161 pKa = 3.91GGAPTACYY169 pKa = 9.86QSCEE173 pKa = 4.11YY174 pKa = 10.76GGNPSPASCYY184 pKa = 9.54LVKK187 pKa = 10.94GSTTTGFCNYY197 pKa = 8.87ILKK200 pKa = 8.92GTGQSCGADD209 pKa = 2.95SYY211 pKa = 11.34TFSQTGDD218 pKa = 3.49SLNPPDD224 pKa = 5.13TPNTDD229 pKa = 3.25PSDD232 pKa = 4.35PNDD235 pKa = 4.31PGCPPGWSWSGTTCVKK251 pKa = 10.28TPTDD255 pKa = 3.83PTDD258 pKa = 3.68PTDD261 pKa = 3.54PTTPGGDD268 pKa = 3.12GGGDD272 pKa = 3.55GNGGGNNNGGGNDD285 pKa = 3.66GGTGNGDD292 pKa = 3.15GSGGGDD298 pKa = 3.31GNGAGDD304 pKa = 4.1GSGDD308 pKa = 3.61GDD310 pKa = 3.91GSGTGGDD317 pKa = 4.04GNGTCDD323 pKa = 3.69PAKK326 pKa = 9.63EE327 pKa = 4.12NCSTGPEE334 pKa = 4.5GPGGEE339 pKa = 4.44LKK341 pKa = 10.79EE342 pKa = 4.34PTPGTWDD349 pKa = 3.43DD350 pKa = 6.49AIATWEE356 pKa = 4.34KK357 pKa = 10.62KK358 pKa = 10.2VEE360 pKa = 4.02EE361 pKa = 4.22AKK363 pKa = 10.76KK364 pKa = 9.88EE365 pKa = 4.02LKK367 pKa = 9.93TKK369 pKa = 10.58VKK371 pKa = 11.07ANVDD375 pKa = 3.5QMKK378 pKa = 10.59GAFDD382 pKa = 4.66LNLAEE387 pKa = 5.82GGGQLPCEE395 pKa = 4.61SMTIWGKK402 pKa = 10.71SYY404 pKa = 10.76SLCISDD410 pKa = 3.97YY411 pKa = 11.17AGQLSSLRR419 pKa = 11.84VALLLMAALIAALILLKK436 pKa = 10.73DD437 pKa = 3.5
MM1 pKa = 7.58SIKK4 pKa = 9.83TLISVLRR11 pKa = 11.84VTLLTACLLPSLFFVRR27 pKa = 11.84SAIAGPYY34 pKa = 7.42IWEE37 pKa = 3.97VVMYY41 pKa = 9.58SSSGSSTPAEE51 pKa = 4.3ACEE54 pKa = 4.04KK55 pKa = 10.65ARR57 pKa = 11.84VVADD61 pKa = 5.44RR62 pKa = 11.84SPDD65 pKa = 3.16WNYY68 pKa = 10.64TSATPKK74 pKa = 10.33MNGLDD79 pKa = 3.38NSYY82 pKa = 10.9CSVVYY87 pKa = 9.5VSRR90 pKa = 11.84RR91 pKa = 11.84DD92 pKa = 3.42PSVVNTCDD100 pKa = 4.67DD101 pKa = 3.73CASWKK106 pKa = 10.32LFRR109 pKa = 11.84KK110 pKa = 10.16GDD112 pKa = 3.54QCANADD118 pKa = 3.63DD119 pKa = 4.74TYY121 pKa = 11.35NASTGICEE129 pKa = 4.52PPPKK133 pKa = 9.72EE134 pKa = 4.2CKK136 pKa = 10.06EE137 pKa = 4.24GEE139 pKa = 4.18LFPAKK144 pKa = 10.51GPDD147 pKa = 3.63SPVVTSGGRR156 pKa = 11.84NYY158 pKa = 10.44VGDD161 pKa = 3.91GGAPTACYY169 pKa = 9.86QSCEE173 pKa = 4.11YY174 pKa = 10.76GGNPSPASCYY184 pKa = 9.54LVKK187 pKa = 10.94GSTTTGFCNYY197 pKa = 8.87ILKK200 pKa = 8.92GTGQSCGADD209 pKa = 2.95SYY211 pKa = 11.34TFSQTGDD218 pKa = 3.49SLNPPDD224 pKa = 5.13TPNTDD229 pKa = 3.25PSDD232 pKa = 4.35PNDD235 pKa = 4.31PGCPPGWSWSGTTCVKK251 pKa = 10.28TPTDD255 pKa = 3.83PTDD258 pKa = 3.68PTDD261 pKa = 3.54PTTPGGDD268 pKa = 3.12GGGDD272 pKa = 3.55GNGGGNNNGGGNDD285 pKa = 3.66GGTGNGDD292 pKa = 3.15GSGGGDD298 pKa = 3.31GNGAGDD304 pKa = 4.1GSGDD308 pKa = 3.61GDD310 pKa = 3.91GSGTGGDD317 pKa = 4.04GNGTCDD323 pKa = 3.69PAKK326 pKa = 9.63EE327 pKa = 4.12NCSTGPEE334 pKa = 4.5GPGGEE339 pKa = 4.44LKK341 pKa = 10.79EE342 pKa = 4.34PTPGTWDD349 pKa = 3.43DD350 pKa = 6.49AIATWEE356 pKa = 4.34KK357 pKa = 10.62KK358 pKa = 10.2VEE360 pKa = 4.02EE361 pKa = 4.22AKK363 pKa = 10.76KK364 pKa = 9.88EE365 pKa = 4.02LKK367 pKa = 9.93TKK369 pKa = 10.58VKK371 pKa = 11.07ANVDD375 pKa = 3.5QMKK378 pKa = 10.59GAFDD382 pKa = 4.66LNLAEE387 pKa = 5.82GGGQLPCEE395 pKa = 4.61SMTIWGKK402 pKa = 10.71SYY404 pKa = 10.76SLCISDD410 pKa = 3.97YY411 pKa = 11.17AGQLSSLRR419 pKa = 11.84VALLLMAALIAALILLKK436 pKa = 10.73DD437 pKa = 3.5
Molecular weight: 44.8 kDa
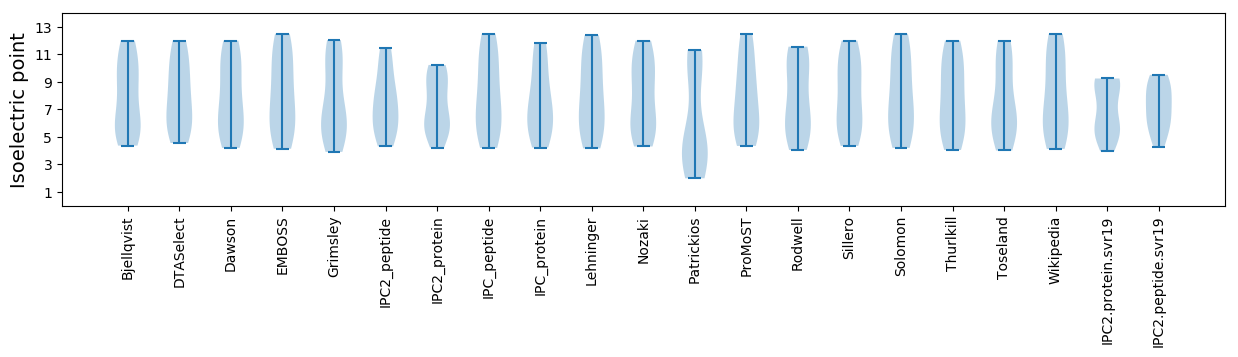
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P25137|VG030_BPPF1 3.2 kDa protein OS=Pseudomonas phage Pf1 OX=2011081 PE=4 SV=1
MM1 pKa = 7.36TKK3 pKa = 10.41YY4 pKa = 10.39LVEE7 pKa = 3.97ICTFHH12 pKa = 7.03GPTRR16 pKa = 11.84QRR18 pKa = 11.84RR19 pKa = 11.84WHH21 pKa = 6.39RR22 pKa = 11.84VHH24 pKa = 6.48QGGSRR29 pKa = 11.84VEE31 pKa = 3.96CQRR34 pKa = 11.84WVEE37 pKa = 3.93EE38 pKa = 4.06LVAVFPTEE46 pKa = 3.8EE47 pKa = 3.73EE48 pKa = 3.86ARR50 pKa = 11.84RR51 pKa = 11.84SFGLTRR57 pKa = 11.84EE58 pKa = 4.32RR59 pKa = 11.84ARR61 pKa = 11.84QVYY64 pKa = 10.08RR65 pKa = 11.84IRR67 pKa = 11.84GVRR70 pKa = 11.84AA71 pKa = 3.04
MM1 pKa = 7.36TKK3 pKa = 10.41YY4 pKa = 10.39LVEE7 pKa = 3.97ICTFHH12 pKa = 7.03GPTRR16 pKa = 11.84QRR18 pKa = 11.84RR19 pKa = 11.84WHH21 pKa = 6.39RR22 pKa = 11.84VHH24 pKa = 6.48QGGSRR29 pKa = 11.84VEE31 pKa = 3.96CQRR34 pKa = 11.84WVEE37 pKa = 3.93EE38 pKa = 4.06LVAVFPTEE46 pKa = 3.8EE47 pKa = 3.73EE48 pKa = 3.86ARR50 pKa = 11.84RR51 pKa = 11.84SFGLTRR57 pKa = 11.84EE58 pKa = 4.32RR59 pKa = 11.84ARR61 pKa = 11.84QVYY64 pKa = 10.08RR65 pKa = 11.84IRR67 pKa = 11.84GVRR70 pKa = 11.84AA71 pKa = 3.04
Molecular weight: 8.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2574 |
30 |
437 |
183.9 |
20.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.868 ± 1.017 | 2.253 ± 0.496 |
5.478 ± 0.697 | 4.74 ± 0.52 |
3.341 ± 0.371 | 9.091 ± 1.207 |
2.214 ± 0.654 | 4.507 ± 0.573 |
4.274 ± 0.367 | 8.936 ± 0.818 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.098 ± 0.214 | 2.681 ± 0.482 |
6.41 ± 0.94 | 3.846 ± 0.549 |
7.265 ± 1.274 | 6.216 ± 0.655 |
5.633 ± 0.657 | 6.061 ± 0.488 |
1.981 ± 0.21 | 3.108 ± 0.323 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
