
Hubei tombus-like virus 9
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.5
Get precalculated fractions of proteins
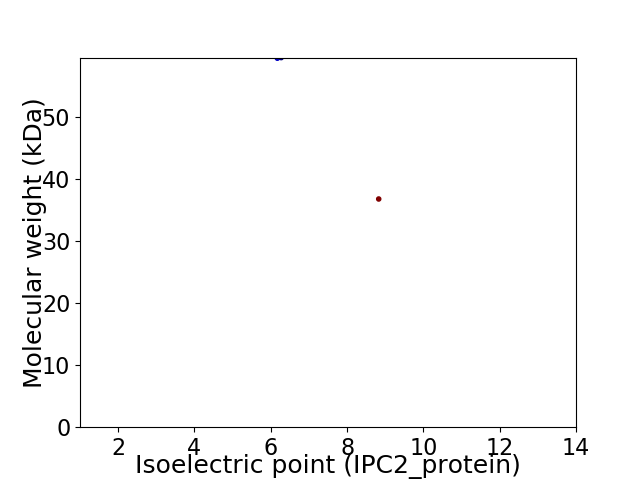
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KGI2|A0A1L3KGI2_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 9 OX=1923296 PE=4 SV=1
MM1 pKa = 7.55VSVQGYY7 pKa = 8.33NSQPKK12 pKa = 8.45FHH14 pKa = 7.33PPGVRR19 pKa = 11.84ISDD22 pKa = 3.74HH23 pKa = 5.79ATSSGRR29 pKa = 11.84EE30 pKa = 4.09KK31 pKa = 10.71NRR33 pKa = 11.84HH34 pKa = 4.47VLVDD38 pKa = 3.24TGVPGVGLCYY48 pKa = 9.62THH50 pKa = 6.93NNSADD55 pKa = 3.39NLLRR59 pKa = 11.84GLGEE63 pKa = 4.2RR64 pKa = 11.84LLMVPTGDD72 pKa = 3.81TFSQPPRR79 pKa = 11.84PCADD83 pKa = 2.89AFRR86 pKa = 11.84CLEE89 pKa = 3.81EE90 pKa = 3.88YY91 pKa = 10.05RR92 pKa = 11.84RR93 pKa = 11.84KK94 pKa = 10.08VVSKK98 pKa = 10.34CPKK101 pKa = 7.71QTGPLEE107 pKa = 4.12RR108 pKa = 11.84DD109 pKa = 3.58EE110 pKa = 5.44FVSLYY115 pKa = 10.66DD116 pKa = 3.29GPKK119 pKa = 8.65RR120 pKa = 11.84KK121 pKa = 9.55RR122 pKa = 11.84YY123 pKa = 8.27EE124 pKa = 3.62AAARR128 pKa = 11.84NLADD132 pKa = 5.92RR133 pKa = 11.84EE134 pKa = 4.22LQKK137 pKa = 11.11ADD139 pKa = 2.62WDD141 pKa = 3.75INVFIKK147 pKa = 10.87DD148 pKa = 3.46EE149 pKa = 4.66TVCSWSKK156 pKa = 10.83VDD158 pKa = 3.7PAPRR162 pKa = 11.84LISPRR167 pKa = 11.84SPEE170 pKa = 3.75YY171 pKa = 10.49CLEE174 pKa = 4.37LGCYY178 pKa = 9.11IKK180 pKa = 10.57PVEE183 pKa = 4.12KK184 pKa = 10.52LLYY187 pKa = 9.96KK188 pKa = 10.56AVARR192 pKa = 11.84VWGEE196 pKa = 3.48VTIAKK201 pKa = 9.76GLNFNQRR208 pKa = 11.84GEE210 pKa = 4.62LIQQKK215 pKa = 8.68WEE217 pKa = 4.18SFTEE221 pKa = 4.3PVAVGLDD228 pKa = 3.21ASRR231 pKa = 11.84FDD233 pKa = 3.5QHH235 pKa = 7.3VSTDD239 pKa = 3.34ALEE242 pKa = 4.53FEE244 pKa = 4.76HH245 pKa = 7.12GFYY248 pKa = 9.29TQLYY252 pKa = 8.09PRR254 pKa = 11.84SKK256 pKa = 10.6KK257 pKa = 10.79LPFLLSKK264 pKa = 10.72QLLNEE269 pKa = 3.41GRR271 pKa = 11.84AYY273 pKa = 11.15VDD275 pKa = 3.54DD276 pKa = 4.79KK277 pKa = 11.28KK278 pKa = 10.87IEE280 pKa = 3.9YY281 pKa = 9.55SVRR284 pKa = 11.84GSRR287 pKa = 11.84MSGDD291 pKa = 3.39MNTALGNCLIMTSLVWGYY309 pKa = 10.56LKK311 pKa = 10.46EE312 pKa = 4.11RR313 pKa = 11.84SVVGKK318 pKa = 9.97LINDD322 pKa = 3.67GDD324 pKa = 4.17DD325 pKa = 3.35CVVIMEE331 pKa = 4.64KK332 pKa = 10.77ADD334 pKa = 3.67LFRR337 pKa = 11.84FMDD340 pKa = 5.15GLTEE344 pKa = 4.19WFTQRR349 pKa = 11.84GFTMKK354 pKa = 10.24VEE356 pKa = 4.37SPVYY360 pKa = 8.5TLEE363 pKa = 4.36EE364 pKa = 3.93IEE366 pKa = 4.77FCQCHH371 pKa = 5.45PVFNGEE377 pKa = 4.36SYY379 pKa = 10.19TMCRR383 pKa = 11.84NVFKK387 pKa = 11.1ALFTDD392 pKa = 4.22VAHH395 pKa = 7.23VGRR398 pKa = 11.84TWRR401 pKa = 11.84EE402 pKa = 3.35VVGIRR407 pKa = 11.84EE408 pKa = 4.44SISVAGAVWAKK419 pKa = 9.86GIPVLSAFYY428 pKa = 10.52RR429 pKa = 11.84SLSTGAPQVVPRR441 pKa = 11.84HH442 pKa = 5.96SGTWWNAQGCNTGTVEE458 pKa = 3.66ITPASRR464 pKa = 11.84EE465 pKa = 4.05SFAKK469 pKa = 10.43AFHH472 pKa = 7.08LDD474 pKa = 3.4PAEE477 pKa = 3.94QAAIEE482 pKa = 4.11QMYY485 pKa = 8.4DD486 pKa = 3.42TLPQLPFSDD495 pKa = 4.37PNRR498 pKa = 11.84MLTYY502 pKa = 10.67NPTIPADD509 pKa = 3.49QYY511 pKa = 10.97PILISEE517 pKa = 4.75PLQVLLFNGPP527 pKa = 3.34
MM1 pKa = 7.55VSVQGYY7 pKa = 8.33NSQPKK12 pKa = 8.45FHH14 pKa = 7.33PPGVRR19 pKa = 11.84ISDD22 pKa = 3.74HH23 pKa = 5.79ATSSGRR29 pKa = 11.84EE30 pKa = 4.09KK31 pKa = 10.71NRR33 pKa = 11.84HH34 pKa = 4.47VLVDD38 pKa = 3.24TGVPGVGLCYY48 pKa = 9.62THH50 pKa = 6.93NNSADD55 pKa = 3.39NLLRR59 pKa = 11.84GLGEE63 pKa = 4.2RR64 pKa = 11.84LLMVPTGDD72 pKa = 3.81TFSQPPRR79 pKa = 11.84PCADD83 pKa = 2.89AFRR86 pKa = 11.84CLEE89 pKa = 3.81EE90 pKa = 3.88YY91 pKa = 10.05RR92 pKa = 11.84RR93 pKa = 11.84KK94 pKa = 10.08VVSKK98 pKa = 10.34CPKK101 pKa = 7.71QTGPLEE107 pKa = 4.12RR108 pKa = 11.84DD109 pKa = 3.58EE110 pKa = 5.44FVSLYY115 pKa = 10.66DD116 pKa = 3.29GPKK119 pKa = 8.65RR120 pKa = 11.84KK121 pKa = 9.55RR122 pKa = 11.84YY123 pKa = 8.27EE124 pKa = 3.62AAARR128 pKa = 11.84NLADD132 pKa = 5.92RR133 pKa = 11.84EE134 pKa = 4.22LQKK137 pKa = 11.11ADD139 pKa = 2.62WDD141 pKa = 3.75INVFIKK147 pKa = 10.87DD148 pKa = 3.46EE149 pKa = 4.66TVCSWSKK156 pKa = 10.83VDD158 pKa = 3.7PAPRR162 pKa = 11.84LISPRR167 pKa = 11.84SPEE170 pKa = 3.75YY171 pKa = 10.49CLEE174 pKa = 4.37LGCYY178 pKa = 9.11IKK180 pKa = 10.57PVEE183 pKa = 4.12KK184 pKa = 10.52LLYY187 pKa = 9.96KK188 pKa = 10.56AVARR192 pKa = 11.84VWGEE196 pKa = 3.48VTIAKK201 pKa = 9.76GLNFNQRR208 pKa = 11.84GEE210 pKa = 4.62LIQQKK215 pKa = 8.68WEE217 pKa = 4.18SFTEE221 pKa = 4.3PVAVGLDD228 pKa = 3.21ASRR231 pKa = 11.84FDD233 pKa = 3.5QHH235 pKa = 7.3VSTDD239 pKa = 3.34ALEE242 pKa = 4.53FEE244 pKa = 4.76HH245 pKa = 7.12GFYY248 pKa = 9.29TQLYY252 pKa = 8.09PRR254 pKa = 11.84SKK256 pKa = 10.6KK257 pKa = 10.79LPFLLSKK264 pKa = 10.72QLLNEE269 pKa = 3.41GRR271 pKa = 11.84AYY273 pKa = 11.15VDD275 pKa = 3.54DD276 pKa = 4.79KK277 pKa = 11.28KK278 pKa = 10.87IEE280 pKa = 3.9YY281 pKa = 9.55SVRR284 pKa = 11.84GSRR287 pKa = 11.84MSGDD291 pKa = 3.39MNTALGNCLIMTSLVWGYY309 pKa = 10.56LKK311 pKa = 10.46EE312 pKa = 4.11RR313 pKa = 11.84SVVGKK318 pKa = 9.97LINDD322 pKa = 3.67GDD324 pKa = 4.17DD325 pKa = 3.35CVVIMEE331 pKa = 4.64KK332 pKa = 10.77ADD334 pKa = 3.67LFRR337 pKa = 11.84FMDD340 pKa = 5.15GLTEE344 pKa = 4.19WFTQRR349 pKa = 11.84GFTMKK354 pKa = 10.24VEE356 pKa = 4.37SPVYY360 pKa = 8.5TLEE363 pKa = 4.36EE364 pKa = 3.93IEE366 pKa = 4.77FCQCHH371 pKa = 5.45PVFNGEE377 pKa = 4.36SYY379 pKa = 10.19TMCRR383 pKa = 11.84NVFKK387 pKa = 11.1ALFTDD392 pKa = 4.22VAHH395 pKa = 7.23VGRR398 pKa = 11.84TWRR401 pKa = 11.84EE402 pKa = 3.35VVGIRR407 pKa = 11.84EE408 pKa = 4.44SISVAGAVWAKK419 pKa = 9.86GIPVLSAFYY428 pKa = 10.52RR429 pKa = 11.84SLSTGAPQVVPRR441 pKa = 11.84HH442 pKa = 5.96SGTWWNAQGCNTGTVEE458 pKa = 3.66ITPASRR464 pKa = 11.84EE465 pKa = 4.05SFAKK469 pKa = 10.43AFHH472 pKa = 7.08LDD474 pKa = 3.4PAEE477 pKa = 3.94QAAIEE482 pKa = 4.11QMYY485 pKa = 8.4DD486 pKa = 3.42TLPQLPFSDD495 pKa = 4.37PNRR498 pKa = 11.84MLTYY502 pKa = 10.67NPTIPADD509 pKa = 3.49QYY511 pKa = 10.97PILISEE517 pKa = 4.75PLQVLLFNGPP527 pKa = 3.34
Molecular weight: 59.41 kDa
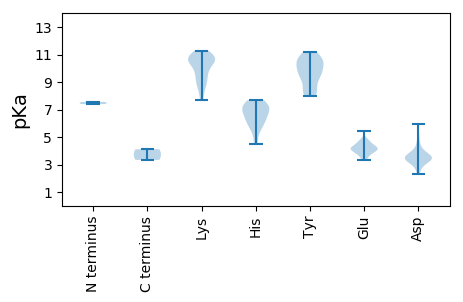
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGI2|A0A1L3KGI2_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 9 OX=1923296 PE=4 SV=1
MM1 pKa = 7.4ARR3 pKa = 11.84KK4 pKa = 9.2KK5 pKa = 10.95QMTKK9 pKa = 9.65PRR11 pKa = 11.84ARR13 pKa = 11.84RR14 pKa = 11.84SRR16 pKa = 11.84KK17 pKa = 8.72QGKK20 pKa = 9.19KK21 pKa = 9.41SSAVPVPKK29 pKa = 10.49RR30 pKa = 11.84MLEE33 pKa = 4.03LAEE36 pKa = 4.94LLHH39 pKa = 6.35NPCDD43 pKa = 3.84ADD45 pKa = 3.48IPRR48 pKa = 11.84GIYY51 pKa = 9.36PGEE54 pKa = 3.96VGAVEE59 pKa = 4.1RR60 pKa = 11.84FIWEE64 pKa = 3.84QAFVGVNNTACFYY77 pKa = 11.01AFHH80 pKa = 7.67PNTGAIIYY88 pKa = 9.77SNQTTSSGALAMTTTASFAPGQTFLSANAAKK119 pKa = 10.44VRR121 pKa = 11.84GLAACIQVACSSLSVTAITGEE142 pKa = 3.67ISLGVLTADD151 pKa = 3.2VVTNNLLTTTTDD163 pKa = 2.34QWFNMLQARR172 pKa = 11.84GPISRR177 pKa = 11.84DD178 pKa = 2.73IKK180 pKa = 9.15EE181 pKa = 4.39VKK183 pKa = 9.02WYY185 pKa = 9.53PGLRR189 pKa = 11.84DD190 pKa = 3.78NSFTTYY196 pKa = 10.57NGSLNSLATLQATGSDD212 pKa = 3.38LSDD215 pKa = 3.13THH217 pKa = 6.27VVCIAIRR224 pKa = 11.84NVPVGTTINVRR235 pKa = 11.84TTYY238 pKa = 9.78VAEE241 pKa = 4.31WSPKK245 pKa = 7.81LTVGITPSTAVSPGTNHH262 pKa = 7.32LAAANILHH270 pKa = 6.62KK271 pKa = 10.82SKK273 pKa = 10.95PNWFHH278 pKa = 6.41NAGSGLGNAASGFATAFMQSVGQNAADD305 pKa = 3.41QTMKK309 pKa = 10.93YY310 pKa = 7.97ITSAGANAGNNMARR324 pKa = 11.84RR325 pKa = 11.84GTVYY329 pKa = 10.49AIEE332 pKa = 4.19EE333 pKa = 4.22LAEE336 pKa = 4.03EE337 pKa = 4.55LGPRR341 pKa = 11.84LLTLL345 pKa = 4.11
MM1 pKa = 7.4ARR3 pKa = 11.84KK4 pKa = 9.2KK5 pKa = 10.95QMTKK9 pKa = 9.65PRR11 pKa = 11.84ARR13 pKa = 11.84RR14 pKa = 11.84SRR16 pKa = 11.84KK17 pKa = 8.72QGKK20 pKa = 9.19KK21 pKa = 9.41SSAVPVPKK29 pKa = 10.49RR30 pKa = 11.84MLEE33 pKa = 4.03LAEE36 pKa = 4.94LLHH39 pKa = 6.35NPCDD43 pKa = 3.84ADD45 pKa = 3.48IPRR48 pKa = 11.84GIYY51 pKa = 9.36PGEE54 pKa = 3.96VGAVEE59 pKa = 4.1RR60 pKa = 11.84FIWEE64 pKa = 3.84QAFVGVNNTACFYY77 pKa = 11.01AFHH80 pKa = 7.67PNTGAIIYY88 pKa = 9.77SNQTTSSGALAMTTTASFAPGQTFLSANAAKK119 pKa = 10.44VRR121 pKa = 11.84GLAACIQVACSSLSVTAITGEE142 pKa = 3.67ISLGVLTADD151 pKa = 3.2VVTNNLLTTTTDD163 pKa = 2.34QWFNMLQARR172 pKa = 11.84GPISRR177 pKa = 11.84DD178 pKa = 2.73IKK180 pKa = 9.15EE181 pKa = 4.39VKK183 pKa = 9.02WYY185 pKa = 9.53PGLRR189 pKa = 11.84DD190 pKa = 3.78NSFTTYY196 pKa = 10.57NGSLNSLATLQATGSDD212 pKa = 3.38LSDD215 pKa = 3.13THH217 pKa = 6.27VVCIAIRR224 pKa = 11.84NVPVGTTINVRR235 pKa = 11.84TTYY238 pKa = 9.78VAEE241 pKa = 4.31WSPKK245 pKa = 7.81LTVGITPSTAVSPGTNHH262 pKa = 7.32LAAANILHH270 pKa = 6.62KK271 pKa = 10.82SKK273 pKa = 10.95PNWFHH278 pKa = 6.41NAGSGLGNAASGFATAFMQSVGQNAADD305 pKa = 3.41QTMKK309 pKa = 10.93YY310 pKa = 7.97ITSAGANAGNNMARR324 pKa = 11.84RR325 pKa = 11.84GTVYY329 pKa = 10.49AIEE332 pKa = 4.19EE333 pKa = 4.22LAEE336 pKa = 4.03EE337 pKa = 4.55LGPRR341 pKa = 11.84LLTLL345 pKa = 4.11
Molecular weight: 36.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
872 |
345 |
527 |
436.0 |
48.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.945 ± 2.162 | 2.064 ± 0.349 |
4.243 ± 0.928 | 5.39 ± 1.085 |
4.014 ± 0.469 | 7.225 ± 0.341 |
1.835 ± 0.054 | 4.243 ± 0.389 |
4.702 ± 0.366 | 8.372 ± 0.31 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.179 ± 0.079 | 4.931 ± 0.985 |
5.734 ± 0.622 | 3.67 ± 0.109 |
5.849 ± 0.523 | 6.995 ± 0.307 |
7.225 ± 1.658 | 7.569 ± 0.512 |
1.72 ± 0.154 | 3.096 ± 0.441 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
