
Rhizobium sp. Leaf306
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Rhizobium; unclassified Rhizobium
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
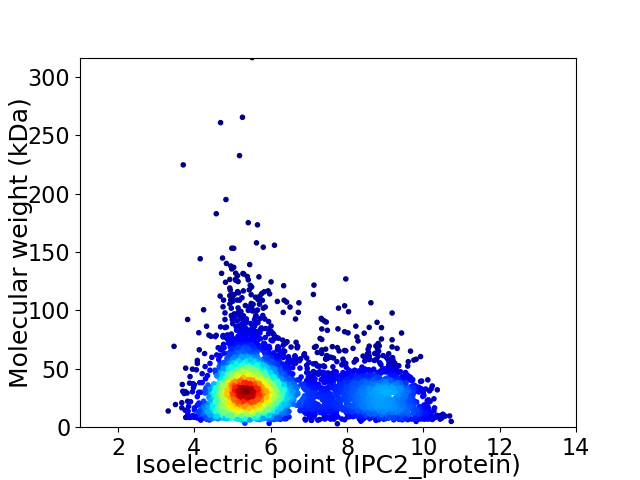
Virtual 2D-PAGE plot for 4551 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
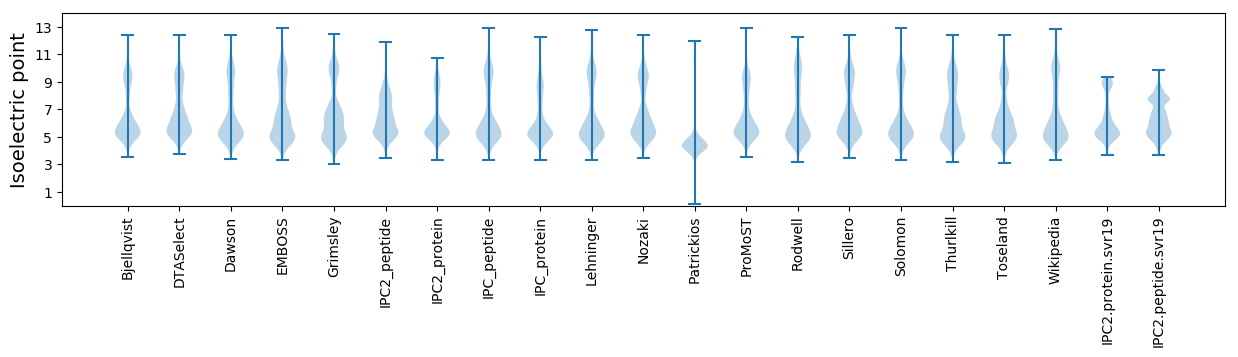
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q5D4H8|A0A0Q5D4H8_9RHIZ Uncharacterized protein OS=Rhizobium sp. Leaf306 OX=1736330 GN=ASG19_20420 PE=4 SV=1
MM1 pKa = 7.46TKK3 pKa = 9.47TVLNALGEE11 pKa = 4.22TLYY14 pKa = 11.38YY15 pKa = 10.75SGTSKK20 pKa = 10.83AWISATGAGTALSGTAANDD39 pKa = 3.32SMYY42 pKa = 11.02GDD44 pKa = 3.71SAVNVTMSGGAGDD57 pKa = 4.08DD58 pKa = 3.44VYY60 pKa = 11.88YY61 pKa = 10.51LYY63 pKa = 11.33SSINRR68 pKa = 11.84AAEE71 pKa = 3.83AAGQGIDD78 pKa = 4.87TVDD81 pKa = 2.44TWMSYY86 pKa = 7.33TLPDD90 pKa = 3.94NIEE93 pKa = 4.04NLRR96 pKa = 11.84VTGDD100 pKa = 2.93NRR102 pKa = 11.84FAFGNDD108 pKa = 2.95LDD110 pKa = 4.95NIISGSSSRR119 pKa = 11.84QTFDD123 pKa = 2.94GGAGNDD129 pKa = 3.49VLTGGGGADD138 pKa = 3.32TFIVAKK144 pKa = 10.77GNGSDD149 pKa = 5.3LITDD153 pKa = 4.3FSADD157 pKa = 3.34DD158 pKa = 4.21KK159 pKa = 11.4IRR161 pKa = 11.84LDD163 pKa = 4.0GYY165 pKa = 10.68SFTSFEE171 pKa = 4.12QVGNSLTQEE180 pKa = 4.29GANLRR185 pKa = 11.84LDD187 pKa = 4.11FGDD190 pKa = 4.03GDD192 pKa = 4.25SLVFAGTTADD202 pKa = 5.46DD203 pKa = 4.21LSADD207 pKa = 3.72QFALSLDD214 pKa = 3.48RR215 pKa = 11.84SVLTKK220 pKa = 10.6TFGDD224 pKa = 3.74EE225 pKa = 4.54FNALSLNNGTSGTWDD240 pKa = 3.87ANFHH244 pKa = 5.85WAPDD248 pKa = 3.67QGSSLPTNGEE258 pKa = 4.13SQWYY262 pKa = 9.11VNPLYY267 pKa = 10.89SPTAGYY273 pKa = 10.89SPFSVSNGALTITAKK288 pKa = 8.95NTPDD292 pKa = 4.65AISDD296 pKa = 3.64AVNGYY301 pKa = 10.8DD302 pKa = 3.97YY303 pKa = 11.62VSGMLNTHH311 pKa = 5.37STFSQTYY318 pKa = 8.57GYY320 pKa = 11.06FEE322 pKa = 4.58MRR324 pKa = 11.84ADD326 pKa = 3.79MPTDD330 pKa = 3.27QATWPAFWLLPEE342 pKa = 5.2DD343 pKa = 4.82GSWPPEE349 pKa = 3.59IDD351 pKa = 3.58VVEE354 pKa = 4.39MRR356 pKa = 11.84GQDD359 pKa = 3.54PNTVHH364 pKa = 6.68VSVHH368 pKa = 5.53SNEE371 pKa = 4.26TGKK374 pKa = 8.36QTTQTSAISVPSTEE388 pKa = 4.56GFHH391 pKa = 6.35TYY393 pKa = 10.26GMLWTEE399 pKa = 4.69EE400 pKa = 4.13EE401 pKa = 4.39IVWYY405 pKa = 10.19FDD407 pKa = 3.49DD408 pKa = 3.92VAIASAEE415 pKa = 4.37TPSDD419 pKa = 3.23MHH421 pKa = 8.3DD422 pKa = 3.32PMYY425 pKa = 11.17LLVNLAVGGAAGKK438 pKa = 9.59PGDD441 pKa = 3.9LSGGAKK447 pKa = 9.03MVIDD451 pKa = 4.49YY452 pKa = 10.67IHH454 pKa = 7.31AYY456 pKa = 9.28EE457 pKa = 4.83INDD460 pKa = 3.67DD461 pKa = 4.02AAPTSSISSASDD473 pKa = 3.21DD474 pKa = 3.76GLVV477 pKa = 3.06
MM1 pKa = 7.46TKK3 pKa = 9.47TVLNALGEE11 pKa = 4.22TLYY14 pKa = 11.38YY15 pKa = 10.75SGTSKK20 pKa = 10.83AWISATGAGTALSGTAANDD39 pKa = 3.32SMYY42 pKa = 11.02GDD44 pKa = 3.71SAVNVTMSGGAGDD57 pKa = 4.08DD58 pKa = 3.44VYY60 pKa = 11.88YY61 pKa = 10.51LYY63 pKa = 11.33SSINRR68 pKa = 11.84AAEE71 pKa = 3.83AAGQGIDD78 pKa = 4.87TVDD81 pKa = 2.44TWMSYY86 pKa = 7.33TLPDD90 pKa = 3.94NIEE93 pKa = 4.04NLRR96 pKa = 11.84VTGDD100 pKa = 2.93NRR102 pKa = 11.84FAFGNDD108 pKa = 2.95LDD110 pKa = 4.95NIISGSSSRR119 pKa = 11.84QTFDD123 pKa = 2.94GGAGNDD129 pKa = 3.49VLTGGGGADD138 pKa = 3.32TFIVAKK144 pKa = 10.77GNGSDD149 pKa = 5.3LITDD153 pKa = 4.3FSADD157 pKa = 3.34DD158 pKa = 4.21KK159 pKa = 11.4IRR161 pKa = 11.84LDD163 pKa = 4.0GYY165 pKa = 10.68SFTSFEE171 pKa = 4.12QVGNSLTQEE180 pKa = 4.29GANLRR185 pKa = 11.84LDD187 pKa = 4.11FGDD190 pKa = 4.03GDD192 pKa = 4.25SLVFAGTTADD202 pKa = 5.46DD203 pKa = 4.21LSADD207 pKa = 3.72QFALSLDD214 pKa = 3.48RR215 pKa = 11.84SVLTKK220 pKa = 10.6TFGDD224 pKa = 3.74EE225 pKa = 4.54FNALSLNNGTSGTWDD240 pKa = 3.87ANFHH244 pKa = 5.85WAPDD248 pKa = 3.67QGSSLPTNGEE258 pKa = 4.13SQWYY262 pKa = 9.11VNPLYY267 pKa = 10.89SPTAGYY273 pKa = 10.89SPFSVSNGALTITAKK288 pKa = 8.95NTPDD292 pKa = 4.65AISDD296 pKa = 3.64AVNGYY301 pKa = 10.8DD302 pKa = 3.97YY303 pKa = 11.62VSGMLNTHH311 pKa = 5.37STFSQTYY318 pKa = 8.57GYY320 pKa = 11.06FEE322 pKa = 4.58MRR324 pKa = 11.84ADD326 pKa = 3.79MPTDD330 pKa = 3.27QATWPAFWLLPEE342 pKa = 5.2DD343 pKa = 4.82GSWPPEE349 pKa = 3.59IDD351 pKa = 3.58VVEE354 pKa = 4.39MRR356 pKa = 11.84GQDD359 pKa = 3.54PNTVHH364 pKa = 6.68VSVHH368 pKa = 5.53SNEE371 pKa = 4.26TGKK374 pKa = 8.36QTTQTSAISVPSTEE388 pKa = 4.56GFHH391 pKa = 6.35TYY393 pKa = 10.26GMLWTEE399 pKa = 4.69EE400 pKa = 4.13EE401 pKa = 4.39IVWYY405 pKa = 10.19FDD407 pKa = 3.49DD408 pKa = 3.92VAIASAEE415 pKa = 4.37TPSDD419 pKa = 3.23MHH421 pKa = 8.3DD422 pKa = 3.32PMYY425 pKa = 11.17LLVNLAVGGAAGKK438 pKa = 9.59PGDD441 pKa = 3.9LSGGAKK447 pKa = 9.03MVIDD451 pKa = 4.49YY452 pKa = 10.67IHH454 pKa = 7.31AYY456 pKa = 9.28EE457 pKa = 4.83INDD460 pKa = 3.67DD461 pKa = 4.02AAPTSSISSASDD473 pKa = 3.21DD474 pKa = 3.76GLVV477 pKa = 3.06
Molecular weight: 50.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q5CRR0|A0A0Q5CRR0_9RHIZ Glutamine amidotransferase OS=Rhizobium sp. Leaf306 OX=1736330 GN=ASG19_16100 PE=3 SV=1
MM1 pKa = 6.95NTVTCHH7 pKa = 4.23GHH9 pKa = 5.63TIRR12 pKa = 11.84MLNISGTIWIAAPDD26 pKa = 3.9ALRR29 pKa = 11.84ALGVDD34 pKa = 3.83IARR37 pKa = 11.84KK38 pKa = 8.37GASVHH43 pKa = 5.26LQKK46 pKa = 10.94LPRR49 pKa = 11.84TEE51 pKa = 3.78VRR53 pKa = 11.84LVTSSQYY60 pKa = 10.77PDD62 pKa = 3.76LFAGRR67 pKa = 11.84RR68 pKa = 11.84GNPRR72 pKa = 11.84MNLVTEE78 pKa = 4.04QGLALLRR85 pKa = 11.84EE86 pKa = 4.28QSGKK90 pKa = 10.24FCAGG94 pKa = 3.09
MM1 pKa = 6.95NTVTCHH7 pKa = 4.23GHH9 pKa = 5.63TIRR12 pKa = 11.84MLNISGTIWIAAPDD26 pKa = 3.9ALRR29 pKa = 11.84ALGVDD34 pKa = 3.83IARR37 pKa = 11.84KK38 pKa = 8.37GASVHH43 pKa = 5.26LQKK46 pKa = 10.94LPRR49 pKa = 11.84TEE51 pKa = 3.78VRR53 pKa = 11.84LVTSSQYY60 pKa = 10.77PDD62 pKa = 3.76LFAGRR67 pKa = 11.84RR68 pKa = 11.84GNPRR72 pKa = 11.84MNLVTEE78 pKa = 4.04QGLALLRR85 pKa = 11.84EE86 pKa = 4.28QSGKK90 pKa = 10.24FCAGG94 pKa = 3.09
Molecular weight: 10.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1439732 |
26 |
2831 |
316.4 |
34.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.562 ± 0.047 | 0.754 ± 0.01 |
5.859 ± 0.031 | 5.696 ± 0.033 |
3.943 ± 0.022 | 8.348 ± 0.031 |
1.948 ± 0.016 | 5.766 ± 0.026 |
3.792 ± 0.03 | 9.884 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.677 ± 0.017 | 2.821 ± 0.019 |
4.849 ± 0.024 | 3.149 ± 0.023 |
6.42 ± 0.032 | 6.074 ± 0.031 |
5.511 ± 0.024 | 7.431 ± 0.028 |
1.229 ± 0.015 | 2.288 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
