
Mongoose feces-associated gemycircularvirus d
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus monas1; Mongoose associated gemycircularvirus 1
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
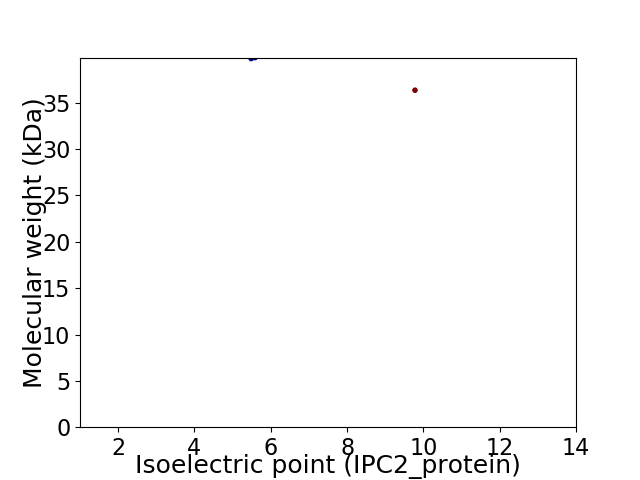
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E3M0E2|A0A0E3M0E2_9VIRU Replication-associated protein OS=Mongoose feces-associated gemycircularvirus d OX=1634487 PE=3 SV=1
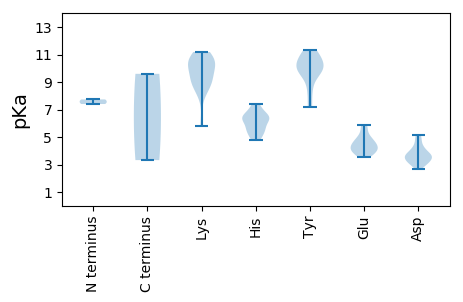
MM1 pKa = 7.4LFVNSRR7 pKa = 11.84YY8 pKa = 10.16LLLTYY13 pKa = 10.06AQSNGLDD20 pKa = 3.1EE21 pKa = 4.66WDD23 pKa = 3.83VSNHH27 pKa = 5.62LSTLGAEE34 pKa = 4.52CIIARR39 pKa = 11.84EE40 pKa = 4.02NHH42 pKa = 6.94DD43 pKa = 4.23DD44 pKa = 3.98GGTHH48 pKa = 6.28LHH50 pKa = 6.56CFVDD54 pKa = 4.87FNRR57 pKa = 11.84KK58 pKa = 8.32FRR60 pKa = 11.84SRR62 pKa = 11.84NVRR65 pKa = 11.84VFDD68 pKa = 3.66VGGFHH73 pKa = 7.41PNISPSRR80 pKa = 11.84GTPEE84 pKa = 3.69KK85 pKa = 10.76GYY87 pKa = 10.89DD88 pKa = 3.47YY89 pKa = 10.69AIKK92 pKa = 10.87DD93 pKa = 3.59GDD95 pKa = 4.09VVAGGLDD102 pKa = 3.28RR103 pKa = 11.84PAPRR107 pKa = 11.84GGMHH111 pKa = 6.28VGAHH115 pKa = 5.22RR116 pKa = 11.84VSNVAHH122 pKa = 6.47LCEE125 pKa = 4.38SSEE128 pKa = 4.2EE129 pKa = 3.9FLEE132 pKa = 5.89LYY134 pKa = 10.95DD135 pKa = 3.74EE136 pKa = 4.52MEE138 pKa = 4.76RR139 pKa = 11.84GDD141 pKa = 4.1LIGRR145 pKa = 11.84FSNVRR150 pKa = 11.84AYY152 pKa = 10.73ADD154 pKa = 2.91WRR156 pKa = 11.84FRR158 pKa = 11.84LDD160 pKa = 2.95IPEE163 pKa = 4.3YY164 pKa = 8.34STPSEE169 pKa = 4.23FVFSSGEE176 pKa = 3.65LDD178 pKa = 5.18GRR180 pKa = 11.84DD181 pKa = 3.29DD182 pKa = 3.61WVQQSGVGSGEE193 pKa = 3.9PLIGMSFTSVRR204 pKa = 11.84LQRR207 pKa = 11.84KK208 pKa = 8.9SLVLYY213 pKa = 10.28GRR215 pKa = 11.84SLTGKK220 pKa = 5.78TTWARR225 pKa = 11.84SLGSHH230 pKa = 6.5MYY232 pKa = 9.84SEE234 pKa = 4.75RR235 pKa = 11.84RR236 pKa = 11.84LNAQMADD243 pKa = 3.72DD244 pKa = 4.51MEE246 pKa = 4.78HH247 pKa = 5.58TAHH250 pKa = 6.19YY251 pKa = 10.15HH252 pKa = 5.36VLDD255 pKa = 4.73DD256 pKa = 3.28VDD258 pKa = 4.94LRR260 pKa = 11.84YY261 pKa = 9.84FPAWKK266 pKa = 9.64SWLGGMQWISNEE278 pKa = 3.51LKK280 pKa = 10.0YY281 pKa = 11.12RR282 pKa = 11.84NVRR285 pKa = 11.84LLKK288 pKa = 9.88WGRR291 pKa = 11.84PCIWCNNTDD300 pKa = 3.18PRR302 pKa = 11.84DD303 pKa = 3.56VMRR306 pKa = 11.84RR307 pKa = 11.84SMAAFDD313 pKa = 3.87GQGDD317 pKa = 4.1GKK319 pKa = 10.74FSHH322 pKa = 7.21EE323 pKa = 3.81DD324 pKa = 3.68LAWLEE329 pKa = 4.13ANCVFIHH336 pKa = 6.53IDD338 pKa = 3.38EE339 pKa = 5.27EE340 pKa = 4.36IATFRR345 pKa = 11.84ASTTT349 pKa = 3.32
MM1 pKa = 7.4LFVNSRR7 pKa = 11.84YY8 pKa = 10.16LLLTYY13 pKa = 10.06AQSNGLDD20 pKa = 3.1EE21 pKa = 4.66WDD23 pKa = 3.83VSNHH27 pKa = 5.62LSTLGAEE34 pKa = 4.52CIIARR39 pKa = 11.84EE40 pKa = 4.02NHH42 pKa = 6.94DD43 pKa = 4.23DD44 pKa = 3.98GGTHH48 pKa = 6.28LHH50 pKa = 6.56CFVDD54 pKa = 4.87FNRR57 pKa = 11.84KK58 pKa = 8.32FRR60 pKa = 11.84SRR62 pKa = 11.84NVRR65 pKa = 11.84VFDD68 pKa = 3.66VGGFHH73 pKa = 7.41PNISPSRR80 pKa = 11.84GTPEE84 pKa = 3.69KK85 pKa = 10.76GYY87 pKa = 10.89DD88 pKa = 3.47YY89 pKa = 10.69AIKK92 pKa = 10.87DD93 pKa = 3.59GDD95 pKa = 4.09VVAGGLDD102 pKa = 3.28RR103 pKa = 11.84PAPRR107 pKa = 11.84GGMHH111 pKa = 6.28VGAHH115 pKa = 5.22RR116 pKa = 11.84VSNVAHH122 pKa = 6.47LCEE125 pKa = 4.38SSEE128 pKa = 4.2EE129 pKa = 3.9FLEE132 pKa = 5.89LYY134 pKa = 10.95DD135 pKa = 3.74EE136 pKa = 4.52MEE138 pKa = 4.76RR139 pKa = 11.84GDD141 pKa = 4.1LIGRR145 pKa = 11.84FSNVRR150 pKa = 11.84AYY152 pKa = 10.73ADD154 pKa = 2.91WRR156 pKa = 11.84FRR158 pKa = 11.84LDD160 pKa = 2.95IPEE163 pKa = 4.3YY164 pKa = 8.34STPSEE169 pKa = 4.23FVFSSGEE176 pKa = 3.65LDD178 pKa = 5.18GRR180 pKa = 11.84DD181 pKa = 3.29DD182 pKa = 3.61WVQQSGVGSGEE193 pKa = 3.9PLIGMSFTSVRR204 pKa = 11.84LQRR207 pKa = 11.84KK208 pKa = 8.9SLVLYY213 pKa = 10.28GRR215 pKa = 11.84SLTGKK220 pKa = 5.78TTWARR225 pKa = 11.84SLGSHH230 pKa = 6.5MYY232 pKa = 9.84SEE234 pKa = 4.75RR235 pKa = 11.84RR236 pKa = 11.84LNAQMADD243 pKa = 3.72DD244 pKa = 4.51MEE246 pKa = 4.78HH247 pKa = 5.58TAHH250 pKa = 6.19YY251 pKa = 10.15HH252 pKa = 5.36VLDD255 pKa = 4.73DD256 pKa = 3.28VDD258 pKa = 4.94LRR260 pKa = 11.84YY261 pKa = 9.84FPAWKK266 pKa = 9.64SWLGGMQWISNEE278 pKa = 3.51LKK280 pKa = 10.0YY281 pKa = 11.12RR282 pKa = 11.84NVRR285 pKa = 11.84LLKK288 pKa = 9.88WGRR291 pKa = 11.84PCIWCNNTDD300 pKa = 3.18PRR302 pKa = 11.84DD303 pKa = 3.56VMRR306 pKa = 11.84RR307 pKa = 11.84SMAAFDD313 pKa = 3.87GQGDD317 pKa = 4.1GKK319 pKa = 10.74FSHH322 pKa = 7.21EE323 pKa = 3.81DD324 pKa = 3.68LAWLEE329 pKa = 4.13ANCVFIHH336 pKa = 6.53IDD338 pKa = 3.38EE339 pKa = 5.27EE340 pKa = 4.36IATFRR345 pKa = 11.84ASTTT349 pKa = 3.32
Molecular weight: 39.78 kDa
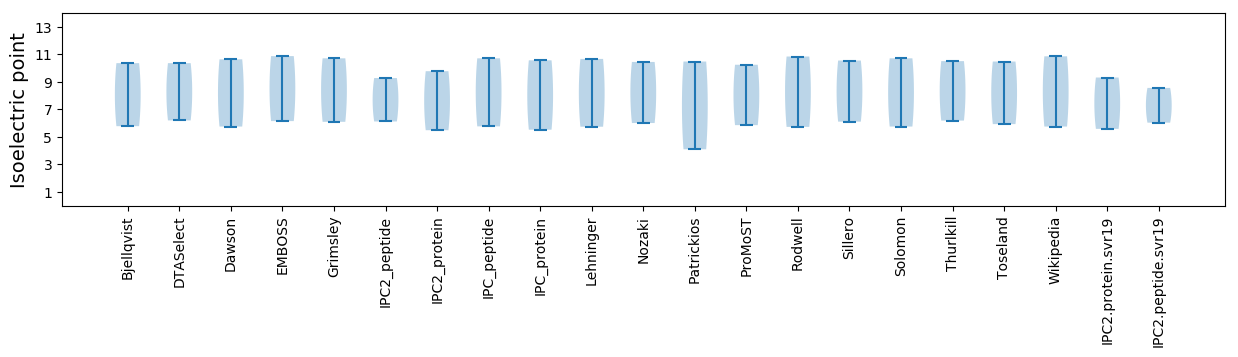
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3M0E2|A0A0E3M0E2_9VIRU Replication-associated protein OS=Mongoose feces-associated gemycircularvirus d OX=1634487 PE=3 SV=1
MM1 pKa = 7.74PPRR4 pKa = 11.84YY5 pKa = 9.21HH6 pKa = 6.29NRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84TIGVSRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.07FTSSRR24 pKa = 11.84YY25 pKa = 8.4RR26 pKa = 11.84KK27 pKa = 9.0PRR29 pKa = 11.84RR30 pKa = 11.84ATRR33 pKa = 11.84RR34 pKa = 11.84AYY36 pKa = 9.88RR37 pKa = 11.84KK38 pKa = 8.87PIRR41 pKa = 11.84RR42 pKa = 11.84VSRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84MINMLSKK54 pKa = 10.66KK55 pKa = 10.3KK56 pKa = 10.0RR57 pKa = 11.84DD58 pKa = 3.6TMLSSAGSGVNPPPDD73 pKa = 3.64STPTGSSIIISPFTSTTNLFEE94 pKa = 4.76SGVHH98 pKa = 5.76SLIFTPSMKK107 pKa = 10.35PLTPSNATFVAQRR120 pKa = 11.84TNTYY124 pKa = 10.12CYY126 pKa = 10.15YY127 pKa = 10.76KK128 pKa = 10.85GFAEE132 pKa = 5.08TYY134 pKa = 9.64TFLPNDD140 pKa = 3.36NSVWWHH146 pKa = 4.82RR147 pKa = 11.84RR148 pKa = 11.84IVFSSRR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84FAEE159 pKa = 3.79LNQITSGSGTLAPAVTAGGISRR181 pKa = 11.84RR182 pKa = 11.84KK183 pKa = 9.16FKK185 pKa = 11.19DD186 pKa = 3.05MSEE189 pKa = 4.13TSGLDD194 pKa = 3.25GFNDD198 pKa = 3.17IQALIIAEE206 pKa = 4.31VFRR209 pKa = 11.84GAYY212 pKa = 7.23NTDD215 pKa = 2.67WADD218 pKa = 3.62PLRR221 pKa = 11.84ARR223 pKa = 11.84LDD225 pKa = 3.43TTKK228 pKa = 10.6INVHH232 pKa = 5.55SDD234 pKa = 2.89RR235 pKa = 11.84LVTLRR240 pKa = 11.84SGNDD244 pKa = 3.01VPAPRR249 pKa = 11.84IVKK252 pKa = 10.0HH253 pKa = 4.85YY254 pKa = 9.66TPINKK259 pKa = 9.01TIVYY263 pKa = 9.39DD264 pKa = 3.79DD265 pKa = 4.21EE266 pKa = 5.1EE267 pKa = 5.73IGVGMQSSYY276 pKa = 11.31YY277 pKa = 10.32AVQGKK282 pKa = 9.03PGIGDD287 pKa = 4.43LYY289 pKa = 11.24VLDD292 pKa = 5.18FFEE295 pKa = 5.71CPVPNDD301 pKa = 3.55TTTTTLNISSQFTSYY316 pKa = 7.21WHH318 pKa = 6.48EE319 pKa = 4.05KK320 pKa = 9.6
MM1 pKa = 7.74PPRR4 pKa = 11.84YY5 pKa = 9.21HH6 pKa = 6.29NRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84TIGVSRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84KK19 pKa = 8.07FTSSRR24 pKa = 11.84YY25 pKa = 8.4RR26 pKa = 11.84KK27 pKa = 9.0PRR29 pKa = 11.84RR30 pKa = 11.84ATRR33 pKa = 11.84RR34 pKa = 11.84AYY36 pKa = 9.88RR37 pKa = 11.84KK38 pKa = 8.87PIRR41 pKa = 11.84RR42 pKa = 11.84VSRR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84MINMLSKK54 pKa = 10.66KK55 pKa = 10.3KK56 pKa = 10.0RR57 pKa = 11.84DD58 pKa = 3.6TMLSSAGSGVNPPPDD73 pKa = 3.64STPTGSSIIISPFTSTTNLFEE94 pKa = 4.76SGVHH98 pKa = 5.76SLIFTPSMKK107 pKa = 10.35PLTPSNATFVAQRR120 pKa = 11.84TNTYY124 pKa = 10.12CYY126 pKa = 10.15YY127 pKa = 10.76KK128 pKa = 10.85GFAEE132 pKa = 5.08TYY134 pKa = 9.64TFLPNDD140 pKa = 3.36NSVWWHH146 pKa = 4.82RR147 pKa = 11.84RR148 pKa = 11.84IVFSSRR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84FAEE159 pKa = 3.79LNQITSGSGTLAPAVTAGGISRR181 pKa = 11.84RR182 pKa = 11.84KK183 pKa = 9.16FKK185 pKa = 11.19DD186 pKa = 3.05MSEE189 pKa = 4.13TSGLDD194 pKa = 3.25GFNDD198 pKa = 3.17IQALIIAEE206 pKa = 4.31VFRR209 pKa = 11.84GAYY212 pKa = 7.23NTDD215 pKa = 2.67WADD218 pKa = 3.62PLRR221 pKa = 11.84ARR223 pKa = 11.84LDD225 pKa = 3.43TTKK228 pKa = 10.6INVHH232 pKa = 5.55SDD234 pKa = 2.89RR235 pKa = 11.84LVTLRR240 pKa = 11.84SGNDD244 pKa = 3.01VPAPRR249 pKa = 11.84IVKK252 pKa = 10.0HH253 pKa = 4.85YY254 pKa = 9.66TPINKK259 pKa = 9.01TIVYY263 pKa = 9.39DD264 pKa = 3.79DD265 pKa = 4.21EE266 pKa = 5.1EE267 pKa = 5.73IGVGMQSSYY276 pKa = 11.31YY277 pKa = 10.32AVQGKK282 pKa = 9.03PGIGDD287 pKa = 4.43LYY289 pKa = 11.24VLDD292 pKa = 5.18FFEE295 pKa = 5.71CPVPNDD301 pKa = 3.55TTTTTLNISSQFTSYY316 pKa = 7.21WHH318 pKa = 6.48EE319 pKa = 4.05KK320 pKa = 9.6
Molecular weight: 36.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
669 |
320 |
349 |
334.5 |
38.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.68 ± 0.264 | 1.196 ± 0.409 |
6.577 ± 1.13 | 4.484 ± 1.198 |
4.783 ± 0.069 | 7.474 ± 1.101 |
2.99 ± 0.799 | 4.783 ± 1.051 |
3.587 ± 0.789 | 7.025 ± 1.228 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.541 ± 0.253 | 4.634 ± 0.263 |
4.783 ± 1.275 | 1.943 ± 0.049 |
9.567 ± 0.759 | 9.118 ± 0.632 |
6.876 ± 2.239 | 5.979 ± 0.254 |
2.093 ± 0.604 | 3.886 ± 0.35 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
