
Changjiang tombus-like virus 22
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 5.44
Get precalculated fractions of proteins
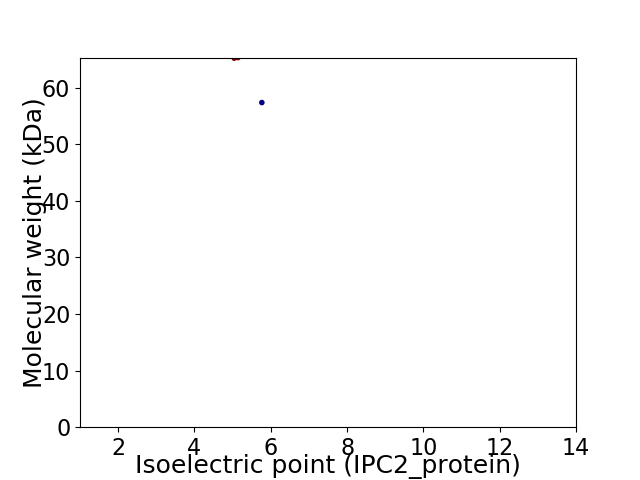
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFX0|A0A1L3KFX0_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 22 OX=1922816 PE=4 SV=1
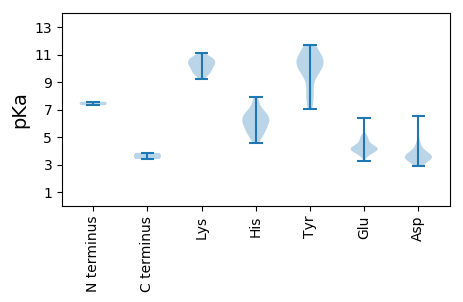
MM1 pKa = 7.57EE2 pKa = 6.37RR3 pKa = 11.84GDD5 pKa = 4.13CDD7 pKa = 5.43LIVPEE12 pKa = 5.02LTADD16 pKa = 3.72DD17 pKa = 4.12NSINRR22 pKa = 11.84PAVGAAFGSFDD33 pKa = 3.62YY34 pKa = 11.72GEE36 pKa = 4.59FSLFSLGLRR45 pKa = 11.84SASRR49 pKa = 11.84LARR52 pKa = 11.84HH53 pKa = 5.98GAVLDD58 pKa = 3.87EE59 pKa = 4.53RR60 pKa = 11.84VGEE63 pKa = 4.28AGSVAGSGDD72 pKa = 3.73LVGDD76 pKa = 3.38SDD78 pKa = 3.76YY79 pKa = 10.91HH80 pKa = 7.94YY81 pKa = 10.69EE82 pKa = 4.16VNSPGWVDD90 pKa = 3.92LSPEE94 pKa = 4.06RR95 pKa = 11.84EE96 pKa = 4.18EE97 pKa = 5.15QSGLDD102 pKa = 3.53PSVPPPAEE110 pKa = 4.13WEE112 pKa = 4.32WVDD115 pKa = 4.26PMSDD119 pKa = 3.56GPDD122 pKa = 3.52GYY124 pKa = 11.55SRR126 pKa = 11.84LPNGEE131 pKa = 4.09IVWNGAEE138 pKa = 4.11NYY140 pKa = 8.28AALMARR146 pKa = 11.84DD147 pKa = 4.63LNHH150 pKa = 6.51EE151 pKa = 4.03MHH153 pKa = 7.1ALCGNRR159 pKa = 11.84PGAPEE164 pKa = 3.92RR165 pKa = 11.84VRR167 pKa = 11.84GRR169 pKa = 11.84GPPLRR174 pKa = 11.84KK175 pKa = 9.42NRR177 pKa = 11.84PRR179 pKa = 11.84VADD182 pKa = 3.72RR183 pKa = 11.84RR184 pKa = 11.84PAVRR188 pKa = 11.84LPPLTRR194 pKa = 11.84EE195 pKa = 3.58GRR197 pKa = 11.84AAVDD201 pKa = 3.53LGFDD205 pKa = 4.95VDD207 pKa = 6.5DD208 pKa = 5.28LDD210 pKa = 3.88QDD212 pKa = 3.54AFEE215 pKa = 5.0FGDD218 pKa = 3.18VGGRR222 pKa = 11.84VGVVEE227 pKa = 4.95RR228 pKa = 11.84GAVIIDD234 pKa = 3.91PPRR237 pKa = 11.84APARR241 pKa = 11.84AFVFGDD247 pKa = 3.02LGEE250 pKa = 4.27PVEE253 pKa = 4.19VEE255 pKa = 3.6EE256 pKa = 5.06GGVVFVNPPRR266 pKa = 11.84CEE268 pKa = 3.91APALPVEE275 pKa = 4.98PIAAKK280 pKa = 10.35GPEE283 pKa = 4.34PEE285 pKa = 4.6VPICLPLPEE294 pKa = 5.69EE295 pKa = 4.2ILEE298 pKa = 4.36STDD301 pKa = 3.21LAAVQGPPLAWDD313 pKa = 3.68VSRR316 pKa = 11.84LVKK319 pKa = 10.03PAFLVPEE326 pKa = 4.44PPDD329 pKa = 3.57NKK331 pKa = 10.54GKK333 pKa = 9.51PRR335 pKa = 11.84DD336 pKa = 3.74LSWDD340 pKa = 3.39EE341 pKa = 3.7RR342 pKa = 11.84VMRR345 pKa = 11.84EE346 pKa = 3.94QKK348 pKa = 10.69LSALGTRR355 pKa = 11.84GSSHH359 pKa = 5.37TAYY362 pKa = 10.78FLGNQTLSMRR372 pKa = 11.84EE373 pKa = 3.5IYY375 pKa = 9.65KK376 pKa = 10.19RR377 pKa = 11.84KK378 pKa = 9.69HH379 pKa = 4.56NRR381 pKa = 11.84RR382 pKa = 11.84WWLWVYY388 pKa = 11.3LLTCYY393 pKa = 10.51LLCSFVFGFSIWLEE407 pKa = 4.32SLWGMVLVSAVVVSAFCLFIWHH429 pKa = 7.53WYY431 pKa = 7.85SVMWGIVTYY440 pKa = 9.45MVVLEE445 pKa = 4.6DD446 pKa = 3.64VPDD449 pKa = 4.21RR450 pKa = 11.84EE451 pKa = 4.54TEE453 pKa = 3.83HH454 pKa = 6.93RR455 pKa = 11.84RR456 pKa = 11.84LADD459 pKa = 3.36SEE461 pKa = 4.27QPDD464 pKa = 3.9IRR466 pKa = 11.84THH468 pKa = 6.4PLAKK472 pKa = 10.12GAIEE476 pKa = 4.1RR477 pKa = 11.84PVKK480 pKa = 9.82EE481 pKa = 3.24RR482 pKa = 11.84RR483 pKa = 11.84ARR485 pKa = 11.84VSRR488 pKa = 11.84VEE490 pKa = 3.84KK491 pKa = 10.6NFWGGEE497 pKa = 4.04FKK499 pKa = 10.62PCRR502 pKa = 11.84DD503 pKa = 2.93RR504 pKa = 11.84TLPVSQQLLDD514 pKa = 3.98DD515 pKa = 4.44CMSPRR520 pKa = 11.84IVPQSIMGDD529 pKa = 3.43DD530 pKa = 3.32AAVAEE535 pKa = 4.32RR536 pKa = 11.84VGRR539 pKa = 11.84YY540 pKa = 8.32LATCSHH546 pKa = 6.46VNTANSDD553 pKa = 3.5QLIQNTGEE561 pKa = 3.97IARR564 pKa = 11.84LLIAEE569 pKa = 5.01RR570 pKa = 11.84FNNWSVHH577 pKa = 5.49RR578 pKa = 11.84GQGGFLPGASS588 pKa = 3.4
MM1 pKa = 7.57EE2 pKa = 6.37RR3 pKa = 11.84GDD5 pKa = 4.13CDD7 pKa = 5.43LIVPEE12 pKa = 5.02LTADD16 pKa = 3.72DD17 pKa = 4.12NSINRR22 pKa = 11.84PAVGAAFGSFDD33 pKa = 3.62YY34 pKa = 11.72GEE36 pKa = 4.59FSLFSLGLRR45 pKa = 11.84SASRR49 pKa = 11.84LARR52 pKa = 11.84HH53 pKa = 5.98GAVLDD58 pKa = 3.87EE59 pKa = 4.53RR60 pKa = 11.84VGEE63 pKa = 4.28AGSVAGSGDD72 pKa = 3.73LVGDD76 pKa = 3.38SDD78 pKa = 3.76YY79 pKa = 10.91HH80 pKa = 7.94YY81 pKa = 10.69EE82 pKa = 4.16VNSPGWVDD90 pKa = 3.92LSPEE94 pKa = 4.06RR95 pKa = 11.84EE96 pKa = 4.18EE97 pKa = 5.15QSGLDD102 pKa = 3.53PSVPPPAEE110 pKa = 4.13WEE112 pKa = 4.32WVDD115 pKa = 4.26PMSDD119 pKa = 3.56GPDD122 pKa = 3.52GYY124 pKa = 11.55SRR126 pKa = 11.84LPNGEE131 pKa = 4.09IVWNGAEE138 pKa = 4.11NYY140 pKa = 8.28AALMARR146 pKa = 11.84DD147 pKa = 4.63LNHH150 pKa = 6.51EE151 pKa = 4.03MHH153 pKa = 7.1ALCGNRR159 pKa = 11.84PGAPEE164 pKa = 3.92RR165 pKa = 11.84VRR167 pKa = 11.84GRR169 pKa = 11.84GPPLRR174 pKa = 11.84KK175 pKa = 9.42NRR177 pKa = 11.84PRR179 pKa = 11.84VADD182 pKa = 3.72RR183 pKa = 11.84RR184 pKa = 11.84PAVRR188 pKa = 11.84LPPLTRR194 pKa = 11.84EE195 pKa = 3.58GRR197 pKa = 11.84AAVDD201 pKa = 3.53LGFDD205 pKa = 4.95VDD207 pKa = 6.5DD208 pKa = 5.28LDD210 pKa = 3.88QDD212 pKa = 3.54AFEE215 pKa = 5.0FGDD218 pKa = 3.18VGGRR222 pKa = 11.84VGVVEE227 pKa = 4.95RR228 pKa = 11.84GAVIIDD234 pKa = 3.91PPRR237 pKa = 11.84APARR241 pKa = 11.84AFVFGDD247 pKa = 3.02LGEE250 pKa = 4.27PVEE253 pKa = 4.19VEE255 pKa = 3.6EE256 pKa = 5.06GGVVFVNPPRR266 pKa = 11.84CEE268 pKa = 3.91APALPVEE275 pKa = 4.98PIAAKK280 pKa = 10.35GPEE283 pKa = 4.34PEE285 pKa = 4.6VPICLPLPEE294 pKa = 5.69EE295 pKa = 4.2ILEE298 pKa = 4.36STDD301 pKa = 3.21LAAVQGPPLAWDD313 pKa = 3.68VSRR316 pKa = 11.84LVKK319 pKa = 10.03PAFLVPEE326 pKa = 4.44PPDD329 pKa = 3.57NKK331 pKa = 10.54GKK333 pKa = 9.51PRR335 pKa = 11.84DD336 pKa = 3.74LSWDD340 pKa = 3.39EE341 pKa = 3.7RR342 pKa = 11.84VMRR345 pKa = 11.84EE346 pKa = 3.94QKK348 pKa = 10.69LSALGTRR355 pKa = 11.84GSSHH359 pKa = 5.37TAYY362 pKa = 10.78FLGNQTLSMRR372 pKa = 11.84EE373 pKa = 3.5IYY375 pKa = 9.65KK376 pKa = 10.19RR377 pKa = 11.84KK378 pKa = 9.69HH379 pKa = 4.56NRR381 pKa = 11.84RR382 pKa = 11.84WWLWVYY388 pKa = 11.3LLTCYY393 pKa = 10.51LLCSFVFGFSIWLEE407 pKa = 4.32SLWGMVLVSAVVVSAFCLFIWHH429 pKa = 7.53WYY431 pKa = 7.85SVMWGIVTYY440 pKa = 9.45MVVLEE445 pKa = 4.6DD446 pKa = 3.64VPDD449 pKa = 4.21RR450 pKa = 11.84EE451 pKa = 4.54TEE453 pKa = 3.83HH454 pKa = 6.93RR455 pKa = 11.84RR456 pKa = 11.84LADD459 pKa = 3.36SEE461 pKa = 4.27QPDD464 pKa = 3.9IRR466 pKa = 11.84THH468 pKa = 6.4PLAKK472 pKa = 10.12GAIEE476 pKa = 4.1RR477 pKa = 11.84PVKK480 pKa = 9.82EE481 pKa = 3.24RR482 pKa = 11.84RR483 pKa = 11.84ARR485 pKa = 11.84VSRR488 pKa = 11.84VEE490 pKa = 3.84KK491 pKa = 10.6NFWGGEE497 pKa = 4.04FKK499 pKa = 10.62PCRR502 pKa = 11.84DD503 pKa = 2.93RR504 pKa = 11.84TLPVSQQLLDD514 pKa = 3.98DD515 pKa = 4.44CMSPRR520 pKa = 11.84IVPQSIMGDD529 pKa = 3.43DD530 pKa = 3.32AAVAEE535 pKa = 4.32RR536 pKa = 11.84VGRR539 pKa = 11.84YY540 pKa = 8.32LATCSHH546 pKa = 6.46VNTANSDD553 pKa = 3.5QLIQNTGEE561 pKa = 3.97IARR564 pKa = 11.84LLIAEE569 pKa = 5.01RR570 pKa = 11.84FNNWSVHH577 pKa = 5.49RR578 pKa = 11.84GQGGFLPGASS588 pKa = 3.4
Molecular weight: 65.2 kDa
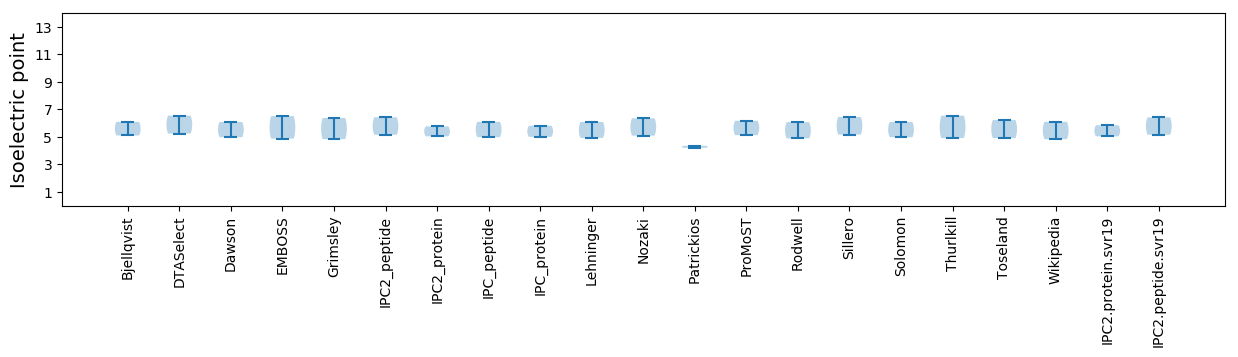
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFX0|A0A1L3KFX0_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 22 OX=1922816 PE=4 SV=1
MM1 pKa = 7.31LFSVGCHH8 pKa = 5.33MDD10 pKa = 3.68GAALPTPDD18 pKa = 3.26ITSSHH23 pKa = 5.92TLLAGVAARR32 pKa = 11.84FATQPPRR39 pKa = 11.84PDD41 pKa = 3.37DD42 pKa = 3.56VLMGEE47 pKa = 4.81LEE49 pKa = 4.21QFVLRR54 pKa = 11.84WLEE57 pKa = 4.02EE58 pKa = 3.78NLTPLSADD66 pKa = 3.51CDD68 pKa = 3.86VSFDD72 pKa = 3.56SWVAGCNQPDD82 pKa = 3.19WRR84 pKa = 11.84KK85 pKa = 10.51AEE87 pKa = 3.9LRR89 pKa = 11.84EE90 pKa = 4.58AFDD93 pKa = 3.26SWVVPEE99 pKa = 4.74SKK101 pKa = 10.72HH102 pKa = 6.39FDD104 pKa = 2.98VKK106 pKa = 10.8MFMKK110 pKa = 10.43KK111 pKa = 9.74EE112 pKa = 4.14SYY114 pKa = 10.18TDD116 pKa = 3.27WKK118 pKa = 9.99HH119 pKa = 5.28GRR121 pKa = 11.84AINSRR126 pKa = 11.84TDD128 pKa = 2.87AFKK131 pKa = 10.87TMVGPFFSAIEE142 pKa = 3.98HH143 pKa = 5.82EE144 pKa = 4.74VFKK147 pKa = 10.95RR148 pKa = 11.84PEE150 pKa = 4.36FIKK153 pKa = 10.5KK154 pKa = 10.18VPVTDD159 pKa = 3.5RR160 pKa = 11.84ASYY163 pKa = 10.26VYY165 pKa = 10.31HH166 pKa = 6.8RR167 pKa = 11.84LYY169 pKa = 9.63RR170 pKa = 11.84TNGHH174 pKa = 6.19YY175 pKa = 10.39YY176 pKa = 10.72SSDD179 pKa = 3.3FTSFEE184 pKa = 3.97ALFTRR189 pKa = 11.84RR190 pKa = 11.84LMEE193 pKa = 3.83IVEE196 pKa = 4.35FSLYY200 pKa = 10.72RR201 pKa = 11.84HH202 pKa = 5.21MTKK205 pKa = 10.24HH206 pKa = 5.91LPSGGWFMGLISRR219 pKa = 11.84VLLGKK224 pKa = 10.24NKK226 pKa = 9.79IKK228 pKa = 10.2NRR230 pKa = 11.84HH231 pKa = 4.9MSLEE235 pKa = 4.13VEE237 pKa = 4.41ATRR240 pKa = 11.84MSGEE244 pKa = 4.07MCTSLGNGFSNLMFNLFVASKK265 pKa = 10.44SGVDD269 pKa = 2.89IDD271 pKa = 4.61GVVEE275 pKa = 4.22GDD277 pKa = 3.89DD278 pKa = 3.88GLFVTSGQIDD288 pKa = 3.54DD289 pKa = 4.49SLFEE293 pKa = 4.18KK294 pKa = 10.69LGLIIKK300 pKa = 9.77IEE302 pKa = 4.0RR303 pKa = 11.84HH304 pKa = 5.86EE305 pKa = 4.82DD306 pKa = 3.07PSTASFCGQIFDD318 pKa = 4.02PQNFVVITDD327 pKa = 3.59PRR329 pKa = 11.84KK330 pKa = 9.44VLASAGWIDD339 pKa = 4.42GKK341 pKa = 11.09YY342 pKa = 10.35LGAKK346 pKa = 9.61RR347 pKa = 11.84GKK349 pKa = 9.74QLGLLRR355 pKa = 11.84AKK357 pKa = 9.94AWSFGYY363 pKa = 10.04QYY365 pKa = 9.82PEE367 pKa = 4.23CPIVSAMARR376 pKa = 11.84ALLRR380 pKa = 11.84LTRR383 pKa = 11.84SMDD386 pKa = 2.92HH387 pKa = 6.25RR388 pKa = 11.84VVFKK392 pKa = 10.73TDD394 pKa = 3.37LGWWLRR400 pKa = 11.84KK401 pKa = 9.22EE402 pKa = 3.79YY403 pKa = 10.34QAAFDD408 pKa = 3.64HH409 pKa = 6.59GRR411 pKa = 11.84PEE413 pKa = 4.0LNRR416 pKa = 11.84PVRR419 pKa = 11.84DD420 pKa = 3.56EE421 pKa = 4.21TRR423 pKa = 11.84SLMHH427 pKa = 6.52KK428 pKa = 10.32VFGVSPDD435 pKa = 3.44TQRR438 pKa = 11.84EE439 pKa = 3.74IEE441 pKa = 3.95AWFDD445 pKa = 3.14NLQEE449 pKa = 4.11IRR451 pKa = 11.84PFPSLDD457 pKa = 3.4VPEE460 pKa = 4.18SWKK463 pKa = 10.71EE464 pKa = 3.74YY465 pKa = 7.62FQSYY469 pKa = 7.06VVRR472 pKa = 11.84TLNTDD477 pKa = 2.89WADD480 pKa = 3.9LPPEE484 pKa = 4.22SWNKK488 pKa = 10.23DD489 pKa = 3.41YY490 pKa = 11.48CVVLPAEE497 pKa = 4.95LDD499 pKa = 3.57PGG501 pKa = 3.84
MM1 pKa = 7.31LFSVGCHH8 pKa = 5.33MDD10 pKa = 3.68GAALPTPDD18 pKa = 3.26ITSSHH23 pKa = 5.92TLLAGVAARR32 pKa = 11.84FATQPPRR39 pKa = 11.84PDD41 pKa = 3.37DD42 pKa = 3.56VLMGEE47 pKa = 4.81LEE49 pKa = 4.21QFVLRR54 pKa = 11.84WLEE57 pKa = 4.02EE58 pKa = 3.78NLTPLSADD66 pKa = 3.51CDD68 pKa = 3.86VSFDD72 pKa = 3.56SWVAGCNQPDD82 pKa = 3.19WRR84 pKa = 11.84KK85 pKa = 10.51AEE87 pKa = 3.9LRR89 pKa = 11.84EE90 pKa = 4.58AFDD93 pKa = 3.26SWVVPEE99 pKa = 4.74SKK101 pKa = 10.72HH102 pKa = 6.39FDD104 pKa = 2.98VKK106 pKa = 10.8MFMKK110 pKa = 10.43KK111 pKa = 9.74EE112 pKa = 4.14SYY114 pKa = 10.18TDD116 pKa = 3.27WKK118 pKa = 9.99HH119 pKa = 5.28GRR121 pKa = 11.84AINSRR126 pKa = 11.84TDD128 pKa = 2.87AFKK131 pKa = 10.87TMVGPFFSAIEE142 pKa = 3.98HH143 pKa = 5.82EE144 pKa = 4.74VFKK147 pKa = 10.95RR148 pKa = 11.84PEE150 pKa = 4.36FIKK153 pKa = 10.5KK154 pKa = 10.18VPVTDD159 pKa = 3.5RR160 pKa = 11.84ASYY163 pKa = 10.26VYY165 pKa = 10.31HH166 pKa = 6.8RR167 pKa = 11.84LYY169 pKa = 9.63RR170 pKa = 11.84TNGHH174 pKa = 6.19YY175 pKa = 10.39YY176 pKa = 10.72SSDD179 pKa = 3.3FTSFEE184 pKa = 3.97ALFTRR189 pKa = 11.84RR190 pKa = 11.84LMEE193 pKa = 3.83IVEE196 pKa = 4.35FSLYY200 pKa = 10.72RR201 pKa = 11.84HH202 pKa = 5.21MTKK205 pKa = 10.24HH206 pKa = 5.91LPSGGWFMGLISRR219 pKa = 11.84VLLGKK224 pKa = 10.24NKK226 pKa = 9.79IKK228 pKa = 10.2NRR230 pKa = 11.84HH231 pKa = 4.9MSLEE235 pKa = 4.13VEE237 pKa = 4.41ATRR240 pKa = 11.84MSGEE244 pKa = 4.07MCTSLGNGFSNLMFNLFVASKK265 pKa = 10.44SGVDD269 pKa = 2.89IDD271 pKa = 4.61GVVEE275 pKa = 4.22GDD277 pKa = 3.89DD278 pKa = 3.88GLFVTSGQIDD288 pKa = 3.54DD289 pKa = 4.49SLFEE293 pKa = 4.18KK294 pKa = 10.69LGLIIKK300 pKa = 9.77IEE302 pKa = 4.0RR303 pKa = 11.84HH304 pKa = 5.86EE305 pKa = 4.82DD306 pKa = 3.07PSTASFCGQIFDD318 pKa = 4.02PQNFVVITDD327 pKa = 3.59PRR329 pKa = 11.84KK330 pKa = 9.44VLASAGWIDD339 pKa = 4.42GKK341 pKa = 11.09YY342 pKa = 10.35LGAKK346 pKa = 9.61RR347 pKa = 11.84GKK349 pKa = 9.74QLGLLRR355 pKa = 11.84AKK357 pKa = 9.94AWSFGYY363 pKa = 10.04QYY365 pKa = 9.82PEE367 pKa = 4.23CPIVSAMARR376 pKa = 11.84ALLRR380 pKa = 11.84LTRR383 pKa = 11.84SMDD386 pKa = 2.92HH387 pKa = 6.25RR388 pKa = 11.84VVFKK392 pKa = 10.73TDD394 pKa = 3.37LGWWLRR400 pKa = 11.84KK401 pKa = 9.22EE402 pKa = 3.79YY403 pKa = 10.34QAAFDD408 pKa = 3.64HH409 pKa = 6.59GRR411 pKa = 11.84PEE413 pKa = 4.0LNRR416 pKa = 11.84PVRR419 pKa = 11.84DD420 pKa = 3.56EE421 pKa = 4.21TRR423 pKa = 11.84SLMHH427 pKa = 6.52KK428 pKa = 10.32VFGVSPDD435 pKa = 3.44TQRR438 pKa = 11.84EE439 pKa = 3.74IEE441 pKa = 3.95AWFDD445 pKa = 3.14NLQEE449 pKa = 4.11IRR451 pKa = 11.84PFPSLDD457 pKa = 3.4VPEE460 pKa = 4.18SWKK463 pKa = 10.71EE464 pKa = 3.74YY465 pKa = 7.62FQSYY469 pKa = 7.06VVRR472 pKa = 11.84TLNTDD477 pKa = 2.89WADD480 pKa = 3.9LPPEE484 pKa = 4.22SWNKK488 pKa = 10.23DD489 pKa = 3.41YY490 pKa = 11.48CVVLPAEE497 pKa = 4.95LDD499 pKa = 3.57PGG501 pKa = 3.84
Molecular weight: 57.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1089 |
501 |
588 |
544.5 |
61.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.346 ± 0.697 | 1.561 ± 0.099 |
6.887 ± 0.06 | 7.071 ± 0.411 |
4.867 ± 0.915 | 7.622 ± 0.743 |
2.296 ± 0.3 | 3.398 ± 0.118 |
3.581 ± 1.088 | 8.999 ± 0.01 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.479 ± 0.43 | 3.03 ± 0.142 |
6.979 ± 0.957 | 2.204 ± 0.115 |
7.805 ± 0.734 | 6.887 ± 0.42 |
3.489 ± 0.783 | 8.356 ± 0.705 |
2.755 ± 0.024 | 2.388 ± 0.245 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
