
Tursiops truncatus papillomavirus 6
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Omikronpapillomavirus; Omikronpapillomavirus 1
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|H6UYQ1|H6UYQ1_PSPV Major capsid protein L1 OS=Tursiops truncatus papillomavirus 6 OX=1144382 GN=L1 PE=3 SV=1
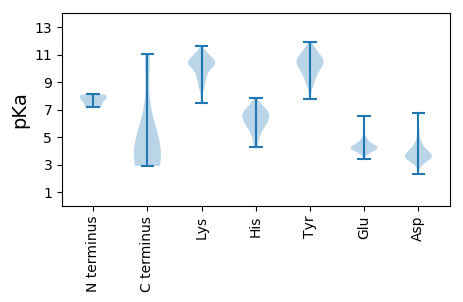
MM1 pKa = 7.23VRR3 pKa = 11.84ATRR6 pKa = 11.84KK7 pKa = 9.58KK8 pKa = 10.11RR9 pKa = 11.84AGEE12 pKa = 3.56QDD14 pKa = 3.88LYY16 pKa = 10.88SGCRR20 pKa = 11.84RR21 pKa = 11.84GQDD24 pKa = 3.32CPDD27 pKa = 4.55DD28 pKa = 3.53IKK30 pKa = 11.62AKK32 pKa = 10.59FEE34 pKa = 3.83QDD36 pKa = 2.48TWADD40 pKa = 3.94RR41 pKa = 11.84LLKK44 pKa = 9.76WFSSFVYY51 pKa = 10.27FGNLGISTGRR61 pKa = 11.84GTGGPSGYY69 pKa = 10.38VPVGSGGRR77 pKa = 11.84GGRR80 pKa = 11.84PAMGGQPSRR89 pKa = 11.84PSLVVEE95 pKa = 4.03NVGPGDD101 pKa = 3.62IPLGGTVDD109 pKa = 3.8ASAPSVITPSEE120 pKa = 4.09STVVVEE126 pKa = 4.95GATTTTEE133 pKa = 4.31EE134 pKa = 4.1IPLVPLHH141 pKa = 7.04PDD143 pKa = 3.39VGPLQPEE150 pKa = 4.4TNIHH154 pKa = 5.25VVPPPEE160 pKa = 5.42AGGPAVLDD168 pKa = 3.44VTSNVTSTYY177 pKa = 9.82PHH179 pKa = 7.48DD180 pKa = 4.34PSIIHH185 pKa = 6.98PGHH188 pKa = 6.4TLLSTYY194 pKa = 10.16INPVFEE200 pKa = 5.79DD201 pKa = 3.76PLFPTVSLQPLDD213 pKa = 3.48TSLIPGEE220 pKa = 4.48TSFPPHH226 pKa = 5.34TVVNLSGTFEE236 pKa = 5.34DD237 pKa = 4.89IEE239 pKa = 5.58LDD241 pKa = 3.82VLDD244 pKa = 5.17GSSKK248 pKa = 10.0LTDD251 pKa = 4.15PKK253 pKa = 10.6TSTPVSRR260 pKa = 11.84VDD262 pKa = 4.48SGLQSIRR269 pKa = 11.84RR270 pKa = 11.84AYY272 pKa = 8.0TRR274 pKa = 11.84RR275 pKa = 11.84ANVLRR280 pKa = 11.84KK281 pKa = 9.39YY282 pKa = 10.48YY283 pKa = 10.95NRR285 pKa = 11.84FTQQVRR291 pKa = 11.84VQKK294 pKa = 10.67PEE296 pKa = 4.27FLTQPSKK303 pKa = 10.91LISYY307 pKa = 7.74EE308 pKa = 3.82FTNQAFDD315 pKa = 4.52PDD317 pKa = 3.79TTLQFPQPADD327 pKa = 3.52EE328 pKa = 4.82VVYY331 pKa = 10.96APDD334 pKa = 4.59NDD336 pKa = 3.93FQDD339 pKa = 3.2VGTLHH344 pKa = 6.75RR345 pKa = 11.84PIYY348 pKa = 9.73SLEE351 pKa = 4.3GGHH354 pKa = 5.9VRR356 pKa = 11.84VSRR359 pKa = 11.84FGVRR363 pKa = 11.84EE364 pKa = 4.06TIRR367 pKa = 11.84TRR369 pKa = 11.84VGTTIGARR377 pKa = 11.84VHH379 pKa = 6.77FFTDD383 pKa = 2.91VSTISNISDD392 pKa = 3.33TFGFTSTLGPDD403 pKa = 3.17VGDD406 pKa = 4.18PGIEE410 pKa = 3.52LHH412 pKa = 7.02LFGEE416 pKa = 4.86STGDD420 pKa = 3.3TSIADD425 pKa = 3.65AQGGGVFLQNGTLQSEE441 pKa = 4.67TQFTDD446 pKa = 3.24VVNGSLQSEE455 pKa = 4.83YY456 pKa = 10.69SDD458 pKa = 4.81SMLLDD463 pKa = 4.12SYY465 pKa = 11.94SEE467 pKa = 4.39TFDD470 pKa = 3.61TAHH473 pKa = 7.33LALLNSNSSSQVLSIPEE490 pKa = 3.94LARR493 pKa = 11.84PFRR496 pKa = 11.84GPAEE500 pKa = 4.38STGGLAVAYY509 pKa = 8.89PVDD512 pKa = 3.75SSLDD516 pKa = 3.36VAAPSTPFIHH526 pKa = 6.43NVPTPPFVVLHH537 pKa = 5.85FSGGGTSFFLHH548 pKa = 6.86PSLLRR553 pKa = 11.84LRR555 pKa = 11.84RR556 pKa = 11.84KK557 pKa = 9.6RR558 pKa = 11.84VFYY561 pKa = 11.01
MM1 pKa = 7.23VRR3 pKa = 11.84ATRR6 pKa = 11.84KK7 pKa = 9.58KK8 pKa = 10.11RR9 pKa = 11.84AGEE12 pKa = 3.56QDD14 pKa = 3.88LYY16 pKa = 10.88SGCRR20 pKa = 11.84RR21 pKa = 11.84GQDD24 pKa = 3.32CPDD27 pKa = 4.55DD28 pKa = 3.53IKK30 pKa = 11.62AKK32 pKa = 10.59FEE34 pKa = 3.83QDD36 pKa = 2.48TWADD40 pKa = 3.94RR41 pKa = 11.84LLKK44 pKa = 9.76WFSSFVYY51 pKa = 10.27FGNLGISTGRR61 pKa = 11.84GTGGPSGYY69 pKa = 10.38VPVGSGGRR77 pKa = 11.84GGRR80 pKa = 11.84PAMGGQPSRR89 pKa = 11.84PSLVVEE95 pKa = 4.03NVGPGDD101 pKa = 3.62IPLGGTVDD109 pKa = 3.8ASAPSVITPSEE120 pKa = 4.09STVVVEE126 pKa = 4.95GATTTTEE133 pKa = 4.31EE134 pKa = 4.1IPLVPLHH141 pKa = 7.04PDD143 pKa = 3.39VGPLQPEE150 pKa = 4.4TNIHH154 pKa = 5.25VVPPPEE160 pKa = 5.42AGGPAVLDD168 pKa = 3.44VTSNVTSTYY177 pKa = 9.82PHH179 pKa = 7.48DD180 pKa = 4.34PSIIHH185 pKa = 6.98PGHH188 pKa = 6.4TLLSTYY194 pKa = 10.16INPVFEE200 pKa = 5.79DD201 pKa = 3.76PLFPTVSLQPLDD213 pKa = 3.48TSLIPGEE220 pKa = 4.48TSFPPHH226 pKa = 5.34TVVNLSGTFEE236 pKa = 5.34DD237 pKa = 4.89IEE239 pKa = 5.58LDD241 pKa = 3.82VLDD244 pKa = 5.17GSSKK248 pKa = 10.0LTDD251 pKa = 4.15PKK253 pKa = 10.6TSTPVSRR260 pKa = 11.84VDD262 pKa = 4.48SGLQSIRR269 pKa = 11.84RR270 pKa = 11.84AYY272 pKa = 8.0TRR274 pKa = 11.84RR275 pKa = 11.84ANVLRR280 pKa = 11.84KK281 pKa = 9.39YY282 pKa = 10.48YY283 pKa = 10.95NRR285 pKa = 11.84FTQQVRR291 pKa = 11.84VQKK294 pKa = 10.67PEE296 pKa = 4.27FLTQPSKK303 pKa = 10.91LISYY307 pKa = 7.74EE308 pKa = 3.82FTNQAFDD315 pKa = 4.52PDD317 pKa = 3.79TTLQFPQPADD327 pKa = 3.52EE328 pKa = 4.82VVYY331 pKa = 10.96APDD334 pKa = 4.59NDD336 pKa = 3.93FQDD339 pKa = 3.2VGTLHH344 pKa = 6.75RR345 pKa = 11.84PIYY348 pKa = 9.73SLEE351 pKa = 4.3GGHH354 pKa = 5.9VRR356 pKa = 11.84VSRR359 pKa = 11.84FGVRR363 pKa = 11.84EE364 pKa = 4.06TIRR367 pKa = 11.84TRR369 pKa = 11.84VGTTIGARR377 pKa = 11.84VHH379 pKa = 6.77FFTDD383 pKa = 2.91VSTISNISDD392 pKa = 3.33TFGFTSTLGPDD403 pKa = 3.17VGDD406 pKa = 4.18PGIEE410 pKa = 3.52LHH412 pKa = 7.02LFGEE416 pKa = 4.86STGDD420 pKa = 3.3TSIADD425 pKa = 3.65AQGGGVFLQNGTLQSEE441 pKa = 4.67TQFTDD446 pKa = 3.24VVNGSLQSEE455 pKa = 4.83YY456 pKa = 10.69SDD458 pKa = 4.81SMLLDD463 pKa = 4.12SYY465 pKa = 11.94SEE467 pKa = 4.39TFDD470 pKa = 3.61TAHH473 pKa = 7.33LALLNSNSSSQVLSIPEE490 pKa = 3.94LARR493 pKa = 11.84PFRR496 pKa = 11.84GPAEE500 pKa = 4.38STGGLAVAYY509 pKa = 8.89PVDD512 pKa = 3.75SSLDD516 pKa = 3.36VAAPSTPFIHH526 pKa = 6.43NVPTPPFVVLHH537 pKa = 5.85FSGGGTSFFLHH548 pKa = 6.86PSLLRR553 pKa = 11.84LRR555 pKa = 11.84RR556 pKa = 11.84KK557 pKa = 9.6RR558 pKa = 11.84VFYY561 pKa = 11.01
Molecular weight: 60.45 kDa
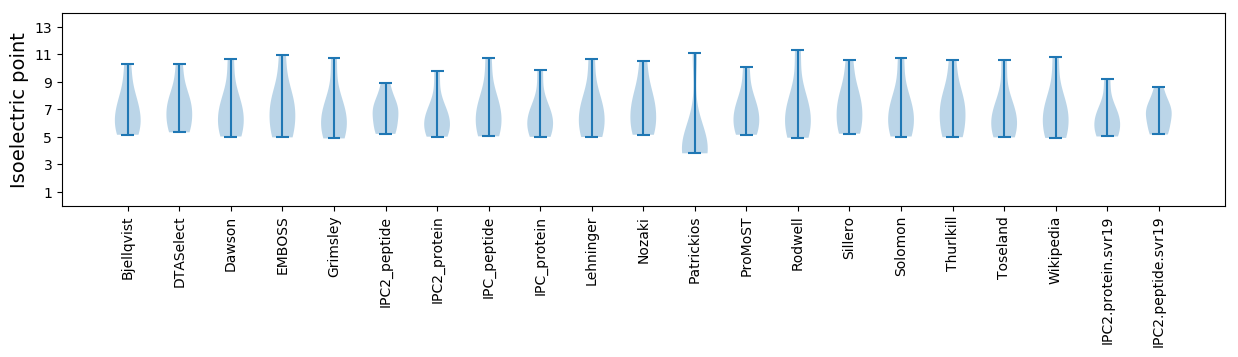
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H6UYQ2|H6UYQ2_PSPV L3 OS=Tursiops truncatus papillomavirus 6 OX=1144382 GN=L3 PE=4 SV=1
MM1 pKa = 8.06EE2 pKa = 4.64IRR4 pKa = 11.84MITTYY9 pKa = 10.68LQQTAKK15 pKa = 9.97KK16 pKa = 8.02WPHH19 pKa = 6.13SFTLQLLVGLLLVVMVSYY37 pKa = 9.17LTGHH41 pKa = 6.27IGFRR45 pKa = 11.84LPRR48 pKa = 11.84VKK50 pKa = 10.04TMAYY54 pKa = 9.27AGEE57 pKa = 4.31MKK59 pKa = 10.37CLSLWLIILEE69 pKa = 4.33TQISLFQLKK78 pKa = 7.79THH80 pKa = 6.9LSMHH84 pKa = 6.58LKK86 pKa = 10.49VPLLQKK92 pKa = 9.33TSKK95 pKa = 8.67FTQGIWRR102 pKa = 11.84NINLISFFSFAKK114 pKa = 10.59SPP116 pKa = 3.37
MM1 pKa = 8.06EE2 pKa = 4.64IRR4 pKa = 11.84MITTYY9 pKa = 10.68LQQTAKK15 pKa = 9.97KK16 pKa = 8.02WPHH19 pKa = 6.13SFTLQLLVGLLLVVMVSYY37 pKa = 9.17LTGHH41 pKa = 6.27IGFRR45 pKa = 11.84LPRR48 pKa = 11.84VKK50 pKa = 10.04TMAYY54 pKa = 9.27AGEE57 pKa = 4.31MKK59 pKa = 10.37CLSLWLIILEE69 pKa = 4.33TQISLFQLKK78 pKa = 7.79THH80 pKa = 6.9LSMHH84 pKa = 6.58LKK86 pKa = 10.49VPLLQKK92 pKa = 9.33TSKK95 pKa = 8.67FTQGIWRR102 pKa = 11.84NINLISFFSFAKK114 pKa = 10.59SPP116 pKa = 3.37
Molecular weight: 13.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2572 |
116 |
642 |
367.4 |
40.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.599 ± 0.488 | 2.216 ± 0.606 |
5.91 ± 0.639 | 5.638 ± 0.524 |
4.471 ± 0.314 | 6.882 ± 0.74 |
2.449 ± 0.456 | 4.199 ± 0.353 |
5.482 ± 0.881 | 8.709 ± 0.646 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.75 ± 0.365 | 3.849 ± 0.402 |
6.493 ± 0.831 | 4.355 ± 0.619 |
4.938 ± 0.243 | 8.981 ± 0.504 |
6.843 ± 0.754 | 6.61 ± 0.66 |
1.711 ± 0.344 | 2.916 ± 0.235 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
