
Prunus avium (Cherry) (Cerasus avium)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae;
Average proteome isoelectric point is 6.64
Get precalculated fractions of proteins
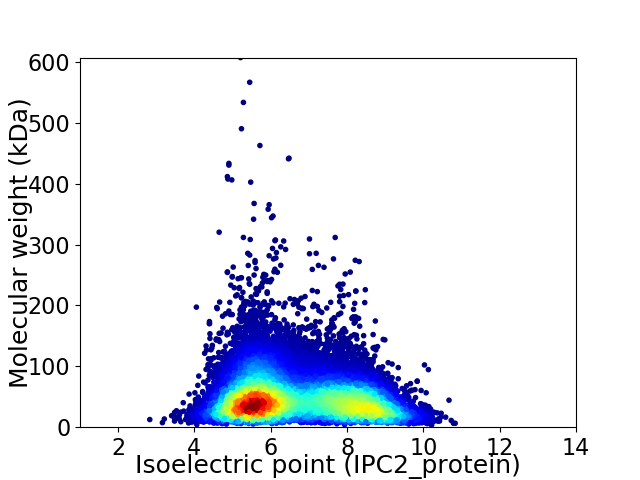
Virtual 2D-PAGE plot for 30515 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P5SZ50|A0A6P5SZ50_PRUAV 30S ribosomal protein 2 chloroplastic OS=Prunus avium OX=42229 GN=LOC110760947 PE=4 SV=1
MM1 pKa = 7.71PSSSSSSPNNYY12 pKa = 9.43NIKK15 pKa = 9.96ILASILFAILLILCAGAGYY34 pKa = 10.92GEE36 pKa = 4.75AQLSPTFYY44 pKa = 11.21DD45 pKa = 3.62EE46 pKa = 5.17DD47 pKa = 4.37CPNATSIVSAVIEE60 pKa = 4.39EE61 pKa = 4.27ALQTDD66 pKa = 3.76PRR68 pKa = 11.84IAASLTRR75 pKa = 11.84LFFHH79 pKa = 7.45DD80 pKa = 4.76CFVNGCDD87 pKa = 5.1GSVLLDD93 pKa = 3.42NSSTIDD99 pKa = 3.49SEE101 pKa = 4.5KK102 pKa = 11.08GAFANNNSARR112 pKa = 11.84GFDD115 pKa = 3.39VVDD118 pKa = 4.33NIKK121 pKa = 8.85TALEE125 pKa = 4.28TACPGIVSCADD136 pKa = 2.91ILAISAEE143 pKa = 4.21EE144 pKa = 4.12SVSLSGGPSWTVLLGRR160 pKa = 11.84RR161 pKa = 11.84DD162 pKa = 3.3STTANRR168 pKa = 11.84TAANEE173 pKa = 3.93ALPAPSFTLDD183 pKa = 3.1EE184 pKa = 4.75LKK186 pKa = 10.98ASFAAVGLDD195 pKa = 3.37TTDD198 pKa = 4.0LVALSGAHH206 pKa = 4.8TFGRR210 pKa = 11.84AQCQFFSDD218 pKa = 3.33RR219 pKa = 11.84LYY221 pKa = 11.26DD222 pKa = 3.96FNSTGSSDD230 pKa = 3.34PTLNSTYY237 pKa = 10.91LEE239 pKa = 4.27TLSALCPEE247 pKa = 4.64SGNGSVLADD256 pKa = 5.1LDD258 pKa = 4.1PSTPDD263 pKa = 3.71VFDD266 pKa = 5.04ADD268 pKa = 4.11YY269 pKa = 10.82FSNLQVHH276 pKa = 6.19YY277 pKa = 10.74GLLQSDD283 pKa = 4.08QEE285 pKa = 4.36LFSTSGADD293 pKa = 3.03TVDD296 pKa = 3.2IVDD299 pKa = 4.51SYY301 pKa = 11.84SANQSAFFEE310 pKa = 4.62SFVISMNKK318 pKa = 8.25MGNISVLTGTDD329 pKa = 3.3GEE331 pKa = 4.36IRR333 pKa = 11.84LNCSKK338 pKa = 11.16VNGDD342 pKa = 4.0SYY344 pKa = 11.66GSSATLIAEE353 pKa = 4.43YY354 pKa = 10.94
MM1 pKa = 7.71PSSSSSSPNNYY12 pKa = 9.43NIKK15 pKa = 9.96ILASILFAILLILCAGAGYY34 pKa = 10.92GEE36 pKa = 4.75AQLSPTFYY44 pKa = 11.21DD45 pKa = 3.62EE46 pKa = 5.17DD47 pKa = 4.37CPNATSIVSAVIEE60 pKa = 4.39EE61 pKa = 4.27ALQTDD66 pKa = 3.76PRR68 pKa = 11.84IAASLTRR75 pKa = 11.84LFFHH79 pKa = 7.45DD80 pKa = 4.76CFVNGCDD87 pKa = 5.1GSVLLDD93 pKa = 3.42NSSTIDD99 pKa = 3.49SEE101 pKa = 4.5KK102 pKa = 11.08GAFANNNSARR112 pKa = 11.84GFDD115 pKa = 3.39VVDD118 pKa = 4.33NIKK121 pKa = 8.85TALEE125 pKa = 4.28TACPGIVSCADD136 pKa = 2.91ILAISAEE143 pKa = 4.21EE144 pKa = 4.12SVSLSGGPSWTVLLGRR160 pKa = 11.84RR161 pKa = 11.84DD162 pKa = 3.3STTANRR168 pKa = 11.84TAANEE173 pKa = 3.93ALPAPSFTLDD183 pKa = 3.1EE184 pKa = 4.75LKK186 pKa = 10.98ASFAAVGLDD195 pKa = 3.37TTDD198 pKa = 4.0LVALSGAHH206 pKa = 4.8TFGRR210 pKa = 11.84AQCQFFSDD218 pKa = 3.33RR219 pKa = 11.84LYY221 pKa = 11.26DD222 pKa = 3.96FNSTGSSDD230 pKa = 3.34PTLNSTYY237 pKa = 10.91LEE239 pKa = 4.27TLSALCPEE247 pKa = 4.64SGNGSVLADD256 pKa = 5.1LDD258 pKa = 4.1PSTPDD263 pKa = 3.71VFDD266 pKa = 5.04ADD268 pKa = 4.11YY269 pKa = 10.82FSNLQVHH276 pKa = 6.19YY277 pKa = 10.74GLLQSDD283 pKa = 4.08QEE285 pKa = 4.36LFSTSGADD293 pKa = 3.03TVDD296 pKa = 3.2IVDD299 pKa = 4.51SYY301 pKa = 11.84SANQSAFFEE310 pKa = 4.62SFVISMNKK318 pKa = 8.25MGNISVLTGTDD329 pKa = 3.3GEE331 pKa = 4.36IRR333 pKa = 11.84LNCSKK338 pKa = 11.16VNGDD342 pKa = 4.0SYY344 pKa = 11.66GSSATLIAEE353 pKa = 4.43YY354 pKa = 10.94
Molecular weight: 37.3 kDa
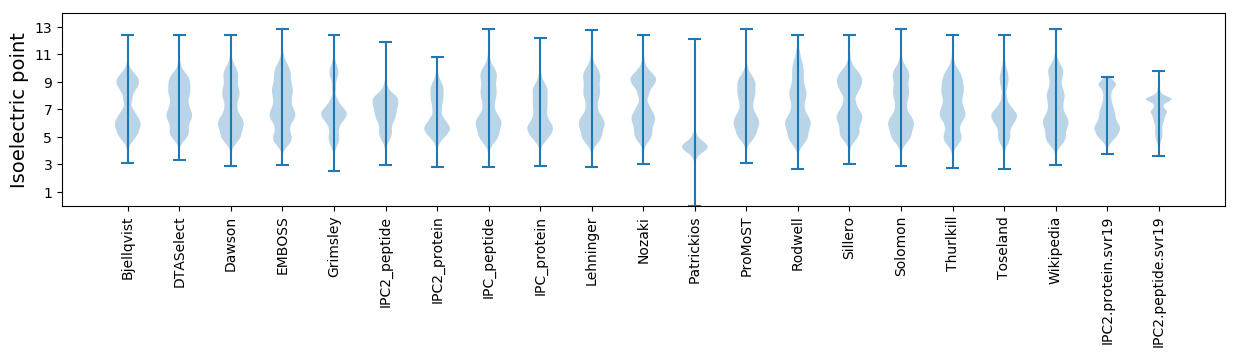
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P5ST86|A0A6P5ST86_PRUAV uncharacterized protein LOC110761795 OS=Prunus avium OX=42229 GN=LOC110761795 PE=4 SV=1
MM1 pKa = 7.71GDD3 pKa = 4.1VISKK7 pKa = 10.35AADD10 pKa = 3.45GVGGLLEE17 pKa = 4.33KK18 pKa = 11.0AFLAPFKK25 pKa = 10.69TMFGGSCEE33 pKa = 4.36DD34 pKa = 3.64VCSGPWDD41 pKa = 4.24FGCFIEE47 pKa = 4.39HH48 pKa = 6.75LCVSSLLKK56 pKa = 10.93LLMILVLCYY65 pKa = 9.14ITLLFFYY72 pKa = 10.9LLFKK76 pKa = 11.01LGLFQCIGKK85 pKa = 8.54SLCKK89 pKa = 9.67MCWAACEE96 pKa = 4.32TYY98 pKa = 9.35WYY100 pKa = 10.14ALEE103 pKa = 6.32DD104 pKa = 3.34ITCFLWYY111 pKa = 9.98KK112 pKa = 10.49LKK114 pKa = 9.08NTKK117 pKa = 9.59RR118 pKa = 11.84VNRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84HH124 pKa = 3.9RR125 pKa = 11.84HH126 pKa = 3.99RR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84SQFRR133 pKa = 11.84DD134 pKa = 2.91IEE136 pKa = 4.17QGYY139 pKa = 10.17SSSEE143 pKa = 4.1EE144 pKa = 3.46NDD146 pKa = 2.87LWDD149 pKa = 4.4NYY151 pKa = 11.08HH152 pKa = 6.59EE153 pKa = 4.95LNVSRR158 pKa = 11.84KK159 pKa = 8.12RR160 pKa = 11.84KK161 pKa = 8.66SVSRR165 pKa = 11.84RR166 pKa = 11.84RR167 pKa = 11.84KK168 pKa = 10.02DD169 pKa = 3.21KK170 pKa = 10.74LQSSVYY176 pKa = 8.36PSSRR180 pKa = 11.84RR181 pKa = 11.84GTHH184 pKa = 4.41SHH186 pKa = 4.68HH187 pKa = 6.47HH188 pKa = 5.74VRR190 pKa = 11.84LKK192 pKa = 10.69SRR194 pKa = 11.84GRR196 pKa = 11.84SVRR199 pKa = 11.84IGGRR203 pKa = 11.84SQRR206 pKa = 11.84PRR208 pKa = 11.84SSRR211 pKa = 11.84HH212 pKa = 3.93LQLRR216 pKa = 11.84KK217 pKa = 9.42VRR219 pKa = 11.84NVRR222 pKa = 11.84RR223 pKa = 11.84QARR226 pKa = 11.84TSKK229 pKa = 10.11RR230 pKa = 11.84RR231 pKa = 11.84KK232 pKa = 9.53LRR234 pKa = 3.61
MM1 pKa = 7.71GDD3 pKa = 4.1VISKK7 pKa = 10.35AADD10 pKa = 3.45GVGGLLEE17 pKa = 4.33KK18 pKa = 11.0AFLAPFKK25 pKa = 10.69TMFGGSCEE33 pKa = 4.36DD34 pKa = 3.64VCSGPWDD41 pKa = 4.24FGCFIEE47 pKa = 4.39HH48 pKa = 6.75LCVSSLLKK56 pKa = 10.93LLMILVLCYY65 pKa = 9.14ITLLFFYY72 pKa = 10.9LLFKK76 pKa = 11.01LGLFQCIGKK85 pKa = 8.54SLCKK89 pKa = 9.67MCWAACEE96 pKa = 4.32TYY98 pKa = 9.35WYY100 pKa = 10.14ALEE103 pKa = 6.32DD104 pKa = 3.34ITCFLWYY111 pKa = 9.98KK112 pKa = 10.49LKK114 pKa = 9.08NTKK117 pKa = 9.59RR118 pKa = 11.84VNRR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84HH124 pKa = 3.9RR125 pKa = 11.84HH126 pKa = 3.99RR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84SQFRR133 pKa = 11.84DD134 pKa = 2.91IEE136 pKa = 4.17QGYY139 pKa = 10.17SSSEE143 pKa = 4.1EE144 pKa = 3.46NDD146 pKa = 2.87LWDD149 pKa = 4.4NYY151 pKa = 11.08HH152 pKa = 6.59EE153 pKa = 4.95LNVSRR158 pKa = 11.84KK159 pKa = 8.12RR160 pKa = 11.84KK161 pKa = 8.66SVSRR165 pKa = 11.84RR166 pKa = 11.84RR167 pKa = 11.84KK168 pKa = 10.02DD169 pKa = 3.21KK170 pKa = 10.74LQSSVYY176 pKa = 8.36PSSRR180 pKa = 11.84RR181 pKa = 11.84GTHH184 pKa = 4.41SHH186 pKa = 4.68HH187 pKa = 6.47HH188 pKa = 5.74VRR190 pKa = 11.84LKK192 pKa = 10.69SRR194 pKa = 11.84GRR196 pKa = 11.84SVRR199 pKa = 11.84IGGRR203 pKa = 11.84SQRR206 pKa = 11.84PRR208 pKa = 11.84SSRR211 pKa = 11.84HH212 pKa = 3.93LQLRR216 pKa = 11.84KK217 pKa = 9.42VRR219 pKa = 11.84NVRR222 pKa = 11.84RR223 pKa = 11.84QARR226 pKa = 11.84TSKK229 pKa = 10.11RR230 pKa = 11.84RR231 pKa = 11.84KK232 pKa = 9.53LRR234 pKa = 3.61
Molecular weight: 27.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
14032431 |
37 |
5357 |
459.9 |
51.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.665 ± 0.013 | 1.884 ± 0.007 |
5.303 ± 0.009 | 6.474 ± 0.015 |
4.201 ± 0.009 | 6.526 ± 0.012 |
2.45 ± 0.006 | 5.247 ± 0.009 |
6.109 ± 0.011 | 9.892 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.366 ± 0.006 | 4.527 ± 0.009 |
4.915 ± 0.013 | 3.795 ± 0.008 |
5.17 ± 0.01 | 8.979 ± 0.015 |
4.891 ± 0.008 | 6.508 ± 0.009 |
1.294 ± 0.005 | 2.798 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
