
Brassica yellows virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; unclassified Polerovirus
Average proteome isoelectric point is 7.44
Get precalculated fractions of proteins
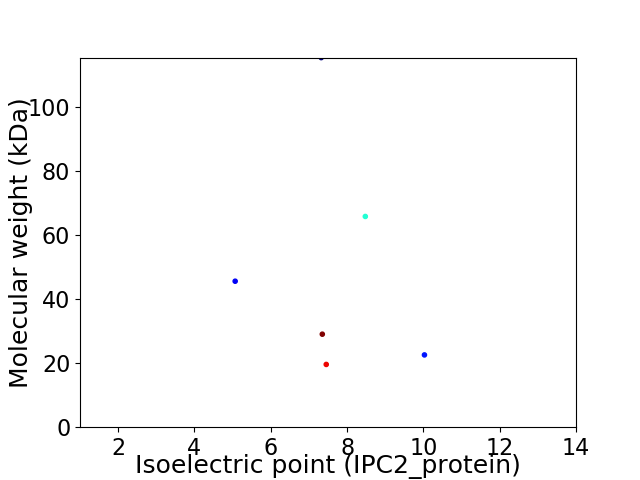
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
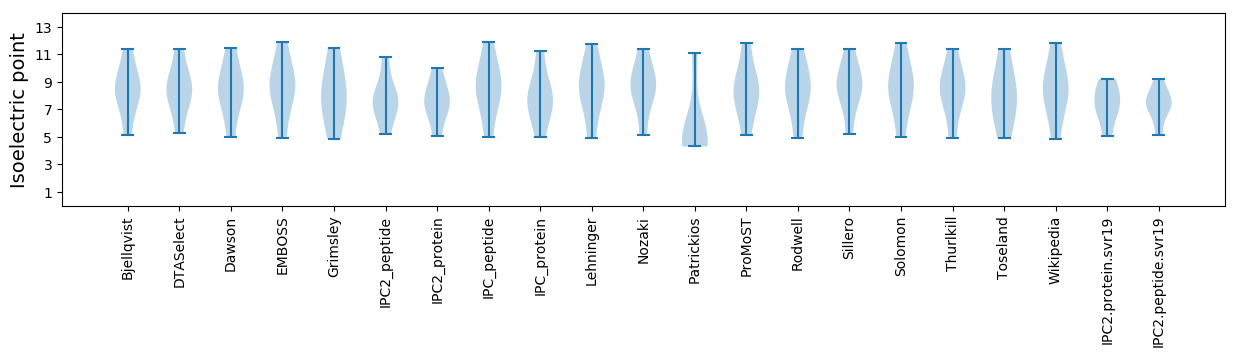
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|G3E231|G3E231_9LUTE Movement protein OS=Brassica yellows virus OX=1046403 PE=3 SV=1
MM1 pKa = 7.33NWTNVDD7 pKa = 3.49ARR9 pKa = 11.84WYY11 pKa = 10.53SSNNVKK17 pKa = 10.16AVPMYY22 pKa = 9.63VFPVPEE28 pKa = 4.42GTWSVEE34 pKa = 3.35ISTEE38 pKa = 4.49GYY40 pKa = 10.03QPTASTTDD48 pKa = 3.66PNKK51 pKa = 10.79GKK53 pKa = 10.38VDD55 pKa = 3.46GMIAYY60 pKa = 10.23SDD62 pKa = 3.96DD63 pKa = 3.63QSEE66 pKa = 4.31VWNVGINQNCNITNLKK82 pKa = 9.98ADD84 pKa = 3.77NSWKK88 pKa = 10.43YY89 pKa = 9.19GHH91 pKa = 7.66PDD93 pKa = 3.15MEE95 pKa = 5.12INNCHH100 pKa = 6.29FNQGQVLEE108 pKa = 4.15MDD110 pKa = 4.0GTISFHH116 pKa = 7.99VEE118 pKa = 3.87TTGSDD123 pKa = 2.85ASFFLVGPAVQKK135 pKa = 9.94QSKK138 pKa = 8.52YY139 pKa = 10.45NYY141 pKa = 8.79AVSYY145 pKa = 8.92GAWTDD150 pKa = 3.01RR151 pKa = 11.84DD152 pKa = 3.83MEE154 pKa = 4.95LGLISVSLDD163 pKa = 3.29EE164 pKa = 4.93KK165 pKa = 11.03DD166 pKa = 3.65GSRR169 pKa = 11.84GSAMKK174 pKa = 10.06RR175 pKa = 11.84PRR177 pKa = 11.84RR178 pKa = 11.84EE179 pKa = 3.83GHH181 pKa = 5.89SKK183 pKa = 10.56AVSTWEE189 pKa = 4.21TINLPEE195 pKa = 4.86KK196 pKa = 10.43EE197 pKa = 4.12NSEE200 pKa = 4.44KK201 pKa = 10.99SITSQRR207 pKa = 11.84QDD209 pKa = 3.05SKK211 pKa = 9.87TQPQQGFSDD220 pKa = 4.45AGSEE224 pKa = 5.01DD225 pKa = 3.42NKK227 pKa = 10.88EE228 pKa = 3.64WDD230 pKa = 3.69FTPGTRR236 pKa = 11.84MFLPEE241 pKa = 4.66DD242 pKa = 3.62FEE244 pKa = 4.77EE245 pKa = 4.5PRR247 pKa = 11.84EE248 pKa = 4.15AQNDD252 pKa = 3.6DD253 pKa = 3.66LLRR256 pKa = 11.84DD257 pKa = 3.56RR258 pKa = 11.84GIVVDD263 pKa = 4.82PSLLEE268 pKa = 4.62IPDD271 pKa = 4.19SPPPRR276 pKa = 11.84ADD278 pKa = 4.79HH279 pKa = 7.36KK280 pKa = 11.17DD281 pKa = 3.21TDD283 pKa = 3.24PWAEE287 pKa = 3.88VEE289 pKa = 4.51RR290 pKa = 11.84YY291 pKa = 9.37KK292 pKa = 10.92SSHH295 pKa = 5.98KK296 pKa = 10.91ALVGEE301 pKa = 4.6DD302 pKa = 3.24TRR304 pKa = 11.84SRR306 pKa = 11.84QSSLTGGSLAGGSLRR321 pKa = 11.84KK322 pKa = 9.71LKK324 pKa = 10.41PVTPDD329 pKa = 2.7EE330 pKa = 4.21YY331 pKa = 11.08EE332 pKa = 3.87QAKK335 pKa = 10.71LKK337 pKa = 10.93DD338 pKa = 3.76EE339 pKa = 4.51SRR341 pKa = 11.84LQYY344 pKa = 10.43EE345 pKa = 3.82QDD347 pKa = 3.0LAEE350 pKa = 4.07YY351 pKa = 9.76HH352 pKa = 6.82RR353 pKa = 11.84SLKK356 pKa = 10.47RR357 pKa = 11.84PSSALIPRR365 pKa = 11.84DD366 pKa = 3.71DD367 pKa = 4.31GEE369 pKa = 4.48LMEE372 pKa = 5.15MDD374 pKa = 4.22PSQRR378 pKa = 11.84KK379 pKa = 8.57LMKK382 pKa = 10.37NAGVNLDD389 pKa = 3.31KK390 pKa = 11.01YY391 pKa = 10.8GRR393 pKa = 11.84PIKK396 pKa = 10.51KK397 pKa = 9.98GFLSFRR403 pKa = 11.84KK404 pKa = 9.7
MM1 pKa = 7.33NWTNVDD7 pKa = 3.49ARR9 pKa = 11.84WYY11 pKa = 10.53SSNNVKK17 pKa = 10.16AVPMYY22 pKa = 9.63VFPVPEE28 pKa = 4.42GTWSVEE34 pKa = 3.35ISTEE38 pKa = 4.49GYY40 pKa = 10.03QPTASTTDD48 pKa = 3.66PNKK51 pKa = 10.79GKK53 pKa = 10.38VDD55 pKa = 3.46GMIAYY60 pKa = 10.23SDD62 pKa = 3.96DD63 pKa = 3.63QSEE66 pKa = 4.31VWNVGINQNCNITNLKK82 pKa = 9.98ADD84 pKa = 3.77NSWKK88 pKa = 10.43YY89 pKa = 9.19GHH91 pKa = 7.66PDD93 pKa = 3.15MEE95 pKa = 5.12INNCHH100 pKa = 6.29FNQGQVLEE108 pKa = 4.15MDD110 pKa = 4.0GTISFHH116 pKa = 7.99VEE118 pKa = 3.87TTGSDD123 pKa = 2.85ASFFLVGPAVQKK135 pKa = 9.94QSKK138 pKa = 8.52YY139 pKa = 10.45NYY141 pKa = 8.79AVSYY145 pKa = 8.92GAWTDD150 pKa = 3.01RR151 pKa = 11.84DD152 pKa = 3.83MEE154 pKa = 4.95LGLISVSLDD163 pKa = 3.29EE164 pKa = 4.93KK165 pKa = 11.03DD166 pKa = 3.65GSRR169 pKa = 11.84GSAMKK174 pKa = 10.06RR175 pKa = 11.84PRR177 pKa = 11.84RR178 pKa = 11.84EE179 pKa = 3.83GHH181 pKa = 5.89SKK183 pKa = 10.56AVSTWEE189 pKa = 4.21TINLPEE195 pKa = 4.86KK196 pKa = 10.43EE197 pKa = 4.12NSEE200 pKa = 4.44KK201 pKa = 10.99SITSQRR207 pKa = 11.84QDD209 pKa = 3.05SKK211 pKa = 9.87TQPQQGFSDD220 pKa = 4.45AGSEE224 pKa = 5.01DD225 pKa = 3.42NKK227 pKa = 10.88EE228 pKa = 3.64WDD230 pKa = 3.69FTPGTRR236 pKa = 11.84MFLPEE241 pKa = 4.66DD242 pKa = 3.62FEE244 pKa = 4.77EE245 pKa = 4.5PRR247 pKa = 11.84EE248 pKa = 4.15AQNDD252 pKa = 3.6DD253 pKa = 3.66LLRR256 pKa = 11.84DD257 pKa = 3.56RR258 pKa = 11.84GIVVDD263 pKa = 4.82PSLLEE268 pKa = 4.62IPDD271 pKa = 4.19SPPPRR276 pKa = 11.84ADD278 pKa = 4.79HH279 pKa = 7.36KK280 pKa = 11.17DD281 pKa = 3.21TDD283 pKa = 3.24PWAEE287 pKa = 3.88VEE289 pKa = 4.51RR290 pKa = 11.84YY291 pKa = 9.37KK292 pKa = 10.92SSHH295 pKa = 5.98KK296 pKa = 10.91ALVGEE301 pKa = 4.6DD302 pKa = 3.24TRR304 pKa = 11.84SRR306 pKa = 11.84QSSLTGGSLAGGSLRR321 pKa = 11.84KK322 pKa = 9.71LKK324 pKa = 10.41PVTPDD329 pKa = 2.7EE330 pKa = 4.21YY331 pKa = 11.08EE332 pKa = 3.87QAKK335 pKa = 10.71LKK337 pKa = 10.93DD338 pKa = 3.76EE339 pKa = 4.51SRR341 pKa = 11.84LQYY344 pKa = 10.43EE345 pKa = 3.82QDD347 pKa = 3.0LAEE350 pKa = 4.07YY351 pKa = 9.76HH352 pKa = 6.82RR353 pKa = 11.84SLKK356 pKa = 10.47RR357 pKa = 11.84PSSALIPRR365 pKa = 11.84DD366 pKa = 3.71DD367 pKa = 4.31GEE369 pKa = 4.48LMEE372 pKa = 5.15MDD374 pKa = 4.22PSQRR378 pKa = 11.84KK379 pKa = 8.57LMKK382 pKa = 10.37NAGVNLDD389 pKa = 3.31KK390 pKa = 11.01YY391 pKa = 10.8GRR393 pKa = 11.84PIKK396 pKa = 10.51KK397 pKa = 9.98GFLSFRR403 pKa = 11.84KK404 pKa = 9.7
Molecular weight: 45.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G3E226|G3E226_9LUTE Read-through protein OS=Brassica yellows virus OX=1046403 PE=4 SV=1
MM1 pKa = 6.49NTVVGRR7 pKa = 11.84RR8 pKa = 11.84TINGRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84QTRR21 pKa = 11.84RR22 pKa = 11.84IQRR25 pKa = 11.84NQPVVVVQASRR36 pKa = 11.84TTQRR40 pKa = 11.84RR41 pKa = 11.84PRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84GNNRR51 pKa = 11.84TGRR54 pKa = 11.84TVPTRR59 pKa = 11.84GAGSSEE65 pKa = 3.84TFVFSKK71 pKa = 11.13DD72 pKa = 3.3NLAGSSSGAITFGPSLSDD90 pKa = 4.1CPAFSNGILKK100 pKa = 10.26AYY102 pKa = 10.04HH103 pKa = 6.58EE104 pKa = 4.78YY105 pKa = 10.01KK106 pKa = 10.07ISMVILEE113 pKa = 4.63FVSEE117 pKa = 4.2ASSQNSGSIAYY128 pKa = 9.39EE129 pKa = 3.85LDD131 pKa = 3.29PHH133 pKa = 6.98CKK135 pKa = 10.11LNSLSSTINKK145 pKa = 9.78FGITKK150 pKa = 9.9PGRR153 pKa = 11.84KK154 pKa = 7.21TFTASFINGIEE165 pKa = 3.93WHH167 pKa = 7.14DD168 pKa = 3.64VAEE171 pKa = 4.32DD172 pKa = 3.49QFRR175 pKa = 11.84ILYY178 pKa = 9.14KK179 pKa = 11.0GNGSSSIAGSFRR191 pKa = 11.84ITMKK195 pKa = 10.75CQFHH199 pKa = 6.24NPKK202 pKa = 10.51
MM1 pKa = 6.49NTVVGRR7 pKa = 11.84RR8 pKa = 11.84TINGRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84QTRR21 pKa = 11.84RR22 pKa = 11.84IQRR25 pKa = 11.84NQPVVVVQASRR36 pKa = 11.84TTQRR40 pKa = 11.84RR41 pKa = 11.84PRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84GNNRR51 pKa = 11.84TGRR54 pKa = 11.84TVPTRR59 pKa = 11.84GAGSSEE65 pKa = 3.84TFVFSKK71 pKa = 11.13DD72 pKa = 3.3NLAGSSSGAITFGPSLSDD90 pKa = 4.1CPAFSNGILKK100 pKa = 10.26AYY102 pKa = 10.04HH103 pKa = 6.58EE104 pKa = 4.78YY105 pKa = 10.01KK106 pKa = 10.07ISMVILEE113 pKa = 4.63FVSEE117 pKa = 4.2ASSQNSGSIAYY128 pKa = 9.39EE129 pKa = 3.85LDD131 pKa = 3.29PHH133 pKa = 6.98CKK135 pKa = 10.11LNSLSSTINKK145 pKa = 9.78FGITKK150 pKa = 9.9PGRR153 pKa = 11.84KK154 pKa = 7.21TFTASFINGIEE165 pKa = 3.93WHH167 pKa = 7.14DD168 pKa = 3.64VAEE171 pKa = 4.32DD172 pKa = 3.49QFRR175 pKa = 11.84ILYY178 pKa = 9.14KK179 pKa = 11.0GNGSSSIAGSFRR191 pKa = 11.84ITMKK195 pKa = 10.75CQFHH199 pKa = 6.24NPKK202 pKa = 10.51
Molecular weight: 22.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2664 |
175 |
1031 |
444.0 |
49.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.719 ± 0.652 | 1.652 ± 0.321 |
4.73 ± 0.768 | 6.006 ± 0.498 |
4.58 ± 0.603 | 6.532 ± 0.27 |
2.14 ± 0.345 | 4.467 ± 0.383 |
5.818 ± 0.381 | 9.197 ± 0.852 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.14 ± 0.176 | 4.429 ± 0.273 |
5.743 ± 0.281 | 3.679 ± 0.462 |
6.194 ± 0.737 | 10.248 ± 0.838 |
5.893 ± 0.378 | 5.33 ± 0.308 |
1.839 ± 0.234 | 2.665 ± 0.215 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
