
Candidatus Sulfobium mesophilum
Taxonomy: cellular organisms; Bacteria; Nitrospirae; Nitrospira; Nitrospirales; Nitrospiraceae; Candidatus Sulfobium
Average proteome isoelectric point is 7.07
Get precalculated fractions of proteins
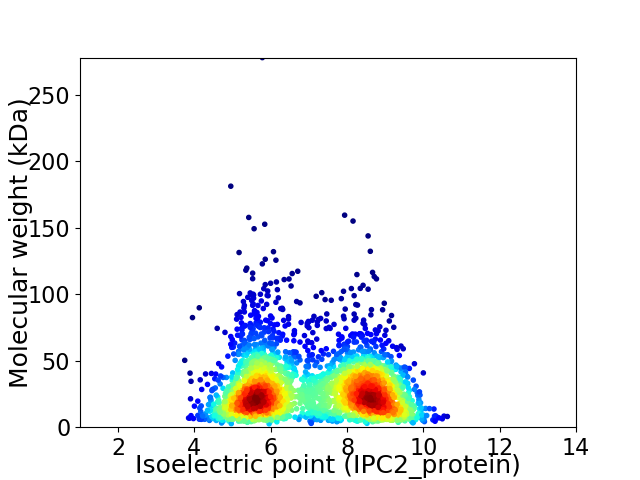
Virtual 2D-PAGE plot for 2739 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2U3QFM9|A0A2U3QFM9_9BACT Uncharacterized protein OS=Candidatus Sulfobium mesophilum OX=2016548 GN=NBG4_190037 PE=4 SV=1
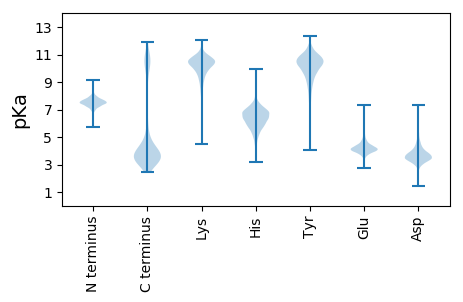
MM1 pKa = 7.51LKK3 pKa = 10.24AKK5 pKa = 10.3KK6 pKa = 10.36SFIVTLFVALVLVSFGIASAASIGGLSSISGPAPVSDD43 pKa = 4.59SYY45 pKa = 12.08VNPDD49 pKa = 2.97GLGDD53 pKa = 3.65VLLFNYY59 pKa = 10.07YY60 pKa = 9.25NARR63 pKa = 11.84SGMEE67 pKa = 3.81TFFAIVNTDD76 pKa = 3.3SEE78 pKa = 4.1QGTRR82 pKa = 11.84VRR84 pKa = 11.84VRR86 pKa = 11.84FRR88 pKa = 11.84EE89 pKa = 3.96AADD92 pKa = 3.31IANSQCVDD100 pKa = 3.01STTGKK105 pKa = 10.44GATAGSFEE113 pKa = 4.13VLDD116 pKa = 4.22FDD118 pKa = 4.47VCLTAGDD125 pKa = 3.68MWSGFVTAAPGGGAMLCSNDD145 pKa = 3.77VDD147 pKa = 3.8TLIYY151 pKa = 10.92DD152 pKa = 4.39GVNSGLFPAGCVPFKK167 pKa = 11.02SGTFGSATLTADD179 pKa = 3.63DD180 pKa = 3.66TMEE183 pKa = 4.9GYY185 pKa = 10.74FEE187 pKa = 5.18VIGEE191 pKa = 4.2DD192 pKa = 3.38SLNAEE197 pKa = 4.45FTTCKK202 pKa = 10.17PIANFQCTGAGTPDD216 pKa = 3.68ACCTAAGLGTCPDD229 pKa = 3.17QFTDD233 pKa = 3.51VGNEE237 pKa = 3.81LFGNAILIDD246 pKa = 3.99ASTSGSWSYY255 pKa = 11.59DD256 pKa = 2.62ATAIADD262 pKa = 3.75FAFEE266 pKa = 5.06SIQQSPTTSRR276 pKa = 11.84PDD278 pKa = 3.32LASDD282 pKa = 3.87SEE284 pKa = 4.5DD285 pKa = 4.1GIDD288 pKa = 4.21GINYY292 pKa = 9.36ILTKK296 pKa = 9.88EE297 pKa = 4.2HH298 pKa = 6.5VFTIYY303 pKa = 11.08DD304 pKa = 4.76LIHH307 pKa = 6.59ARR309 pKa = 11.84TEE311 pKa = 3.96IVLTEE316 pKa = 3.93PTKK319 pKa = 11.07LLTQQNVGCSLEE331 pKa = 4.05IPGTFNPNIFTDD343 pKa = 4.25DD344 pKa = 3.53SALFTVWNDD353 pKa = 3.2SEE355 pKa = 4.5KK356 pKa = 10.83QLTTVCQFSPCPGGTVNKK374 pKa = 9.94LPHH377 pKa = 6.28EE378 pKa = 4.51VNVLQMNIPTAGVVSDD394 pKa = 4.3ILDD397 pKa = 3.62SAVDD401 pKa = 3.48QVVAITFDD409 pKa = 3.81FGWIDD414 pKa = 3.35INLDD418 pKa = 3.27PDD420 pKa = 3.77VNLTFPHH427 pKa = 5.09QTCFGDD433 pKa = 4.75DD434 pKa = 3.01NVAPFNCSNGWPVLGITLLDD454 pKa = 3.64VDD456 pKa = 5.01HH457 pKa = 6.75GASTGAFPTQYY468 pKa = 9.32FTDD471 pKa = 3.63AFLDD475 pKa = 3.67
MM1 pKa = 7.51LKK3 pKa = 10.24AKK5 pKa = 10.3KK6 pKa = 10.36SFIVTLFVALVLVSFGIASAASIGGLSSISGPAPVSDD43 pKa = 4.59SYY45 pKa = 12.08VNPDD49 pKa = 2.97GLGDD53 pKa = 3.65VLLFNYY59 pKa = 10.07YY60 pKa = 9.25NARR63 pKa = 11.84SGMEE67 pKa = 3.81TFFAIVNTDD76 pKa = 3.3SEE78 pKa = 4.1QGTRR82 pKa = 11.84VRR84 pKa = 11.84VRR86 pKa = 11.84FRR88 pKa = 11.84EE89 pKa = 3.96AADD92 pKa = 3.31IANSQCVDD100 pKa = 3.01STTGKK105 pKa = 10.44GATAGSFEE113 pKa = 4.13VLDD116 pKa = 4.22FDD118 pKa = 4.47VCLTAGDD125 pKa = 3.68MWSGFVTAAPGGGAMLCSNDD145 pKa = 3.77VDD147 pKa = 3.8TLIYY151 pKa = 10.92DD152 pKa = 4.39GVNSGLFPAGCVPFKK167 pKa = 11.02SGTFGSATLTADD179 pKa = 3.63DD180 pKa = 3.66TMEE183 pKa = 4.9GYY185 pKa = 10.74FEE187 pKa = 5.18VIGEE191 pKa = 4.2DD192 pKa = 3.38SLNAEE197 pKa = 4.45FTTCKK202 pKa = 10.17PIANFQCTGAGTPDD216 pKa = 3.68ACCTAAGLGTCPDD229 pKa = 3.17QFTDD233 pKa = 3.51VGNEE237 pKa = 3.81LFGNAILIDD246 pKa = 3.99ASTSGSWSYY255 pKa = 11.59DD256 pKa = 2.62ATAIADD262 pKa = 3.75FAFEE266 pKa = 5.06SIQQSPTTSRR276 pKa = 11.84PDD278 pKa = 3.32LASDD282 pKa = 3.87SEE284 pKa = 4.5DD285 pKa = 4.1GIDD288 pKa = 4.21GINYY292 pKa = 9.36ILTKK296 pKa = 9.88EE297 pKa = 4.2HH298 pKa = 6.5VFTIYY303 pKa = 11.08DD304 pKa = 4.76LIHH307 pKa = 6.59ARR309 pKa = 11.84TEE311 pKa = 3.96IVLTEE316 pKa = 3.93PTKK319 pKa = 11.07LLTQQNVGCSLEE331 pKa = 4.05IPGTFNPNIFTDD343 pKa = 4.25DD344 pKa = 3.53SALFTVWNDD353 pKa = 3.2SEE355 pKa = 4.5KK356 pKa = 10.83QLTTVCQFSPCPGGTVNKK374 pKa = 9.94LPHH377 pKa = 6.28EE378 pKa = 4.51VNVLQMNIPTAGVVSDD394 pKa = 4.3ILDD397 pKa = 3.62SAVDD401 pKa = 3.48QVVAITFDD409 pKa = 3.81FGWIDD414 pKa = 3.35INLDD418 pKa = 3.27PDD420 pKa = 3.77VNLTFPHH427 pKa = 5.09QTCFGDD433 pKa = 4.75DD434 pKa = 3.01NVAPFNCSNGWPVLGITLLDD454 pKa = 3.64VDD456 pKa = 5.01HH457 pKa = 6.75GASTGAFPTQYY468 pKa = 9.32FTDD471 pKa = 3.63AFLDD475 pKa = 3.67
Molecular weight: 50.27 kDa
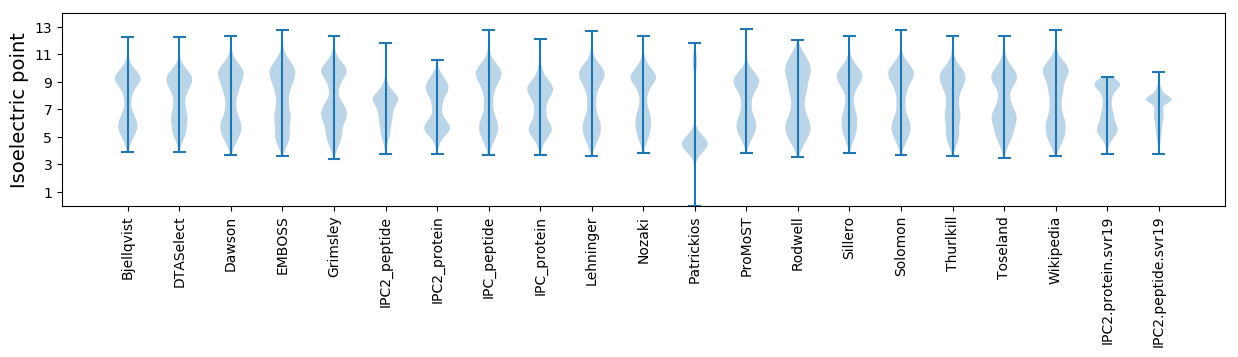
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U3QKZ1|A0A2U3QKZ1_9BACT Putative streptogramin lyase gluconolactonase family protein OS=Candidatus Sulfobium mesophilum OX=2016548 GN=NBG4_90020 PE=4 SV=1
MM1 pKa = 7.64GKK3 pKa = 10.04NNFWSPYY10 pKa = 7.76IAGIGLGLTLLATFYY25 pKa = 11.05IMGKK29 pKa = 10.24GLGASSAFSIVSAVAVEE46 pKa = 4.44KK47 pKa = 10.73INPEE51 pKa = 3.55FAGNLRR57 pKa = 11.84YY58 pKa = 9.57FSRR61 pKa = 11.84YY62 pKa = 9.64LSTASPLKK70 pKa = 10.21DD71 pKa = 2.99WIVFEE76 pKa = 4.3ILGLFAGALTGALLSRR92 pKa = 11.84NFQIRR97 pKa = 11.84FDD99 pKa = 4.02KK100 pKa = 10.9APGMGGTTRR109 pKa = 11.84LFTAFTGGILIGFAGRR125 pKa = 11.84LARR128 pKa = 11.84GCTSGVALSGGAQLAAAGWIFVISMFISGFLFAAIFRR165 pKa = 11.84RR166 pKa = 11.84LWSS169 pKa = 3.48
MM1 pKa = 7.64GKK3 pKa = 10.04NNFWSPYY10 pKa = 7.76IAGIGLGLTLLATFYY25 pKa = 11.05IMGKK29 pKa = 10.24GLGASSAFSIVSAVAVEE46 pKa = 4.44KK47 pKa = 10.73INPEE51 pKa = 3.55FAGNLRR57 pKa = 11.84YY58 pKa = 9.57FSRR61 pKa = 11.84YY62 pKa = 9.64LSTASPLKK70 pKa = 10.21DD71 pKa = 2.99WIVFEE76 pKa = 4.3ILGLFAGALTGALLSRR92 pKa = 11.84NFQIRR97 pKa = 11.84FDD99 pKa = 4.02KK100 pKa = 10.9APGMGGTTRR109 pKa = 11.84LFTAFTGGILIGFAGRR125 pKa = 11.84LARR128 pKa = 11.84GCTSGVALSGGAQLAAAGWIFVISMFISGFLFAAIFRR165 pKa = 11.84RR166 pKa = 11.84LWSS169 pKa = 3.48
Molecular weight: 17.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
780115 |
22 |
2513 |
284.8 |
31.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.979 ± 0.051 | 1.133 ± 0.022 |
5.238 ± 0.034 | 6.732 ± 0.058 |
4.457 ± 0.039 | 7.748 ± 0.048 |
1.894 ± 0.021 | 7.271 ± 0.04 |
6.675 ± 0.052 | 9.684 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.718 ± 0.022 | 3.442 ± 0.026 |
4.089 ± 0.03 | 2.558 ± 0.026 |
5.751 ± 0.043 | 6.387 ± 0.036 |
5.013 ± 0.036 | 7.145 ± 0.042 |
0.977 ± 0.019 | 3.109 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
