
Capybara microvirus Cap3_SP_465
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.45
Get precalculated fractions of proteins
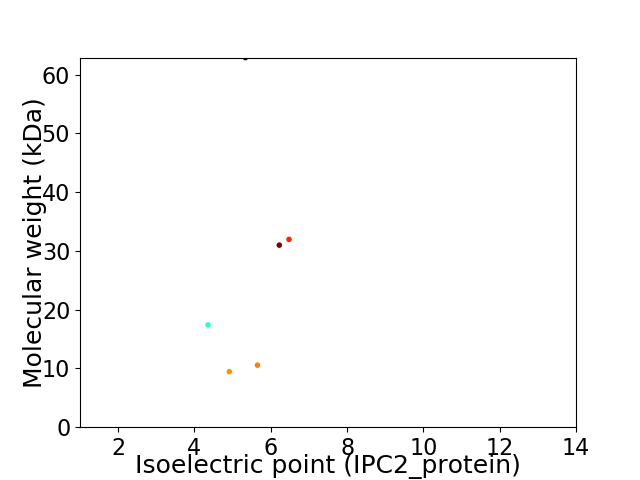
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W588|A0A4P8W588_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_465 OX=2585462 PE=3 SV=1
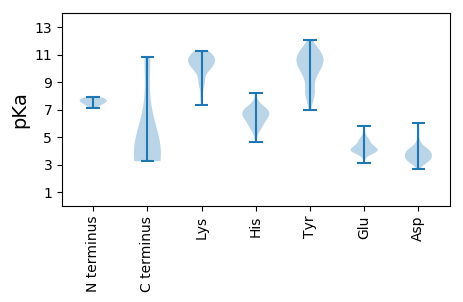
MM1 pKa = 7.54KK2 pKa = 10.18IEE4 pKa = 4.5FKK6 pKa = 11.13NPLEE10 pKa = 4.96IDD12 pKa = 3.27TSSYY16 pKa = 11.39VSVCGLPYY24 pKa = 10.61EE25 pKa = 5.4DD26 pKa = 6.06DD27 pKa = 3.99YY28 pKa = 12.05QLSLDD33 pKa = 4.53DD34 pKa = 5.3NGCEE38 pKa = 3.96YY39 pKa = 10.71LKK41 pKa = 10.74KK42 pKa = 10.62VGVIEE47 pKa = 4.38TYY49 pKa = 11.14KK50 pKa = 10.65LIQSHH55 pKa = 6.97RR56 pKa = 11.84DD57 pKa = 3.42SCDD60 pKa = 2.78LAYY63 pKa = 9.8ILDD66 pKa = 4.04TLDD69 pKa = 4.08PDD71 pKa = 4.01SVLGSGSPNVSFDD84 pKa = 4.64DD85 pKa = 4.14IYY87 pKa = 11.05IDD89 pKa = 3.7FTKK92 pKa = 10.63FPTNPGEE99 pKa = 4.13AFNLIKK105 pKa = 10.72NAEE108 pKa = 3.99FAFNKK113 pKa = 10.34LPFEE117 pKa = 4.74LRR119 pKa = 11.84QEE121 pKa = 4.75CNFSPNVFVKK131 pKa = 10.76NLDD134 pKa = 3.35KK135 pKa = 11.04LAEE138 pKa = 4.32KK139 pKa = 10.31FSKK142 pKa = 9.87VDD144 pKa = 3.12KK145 pKa = 10.5VEE147 pKa = 4.2SVGVSNEE154 pKa = 3.56
MM1 pKa = 7.54KK2 pKa = 10.18IEE4 pKa = 4.5FKK6 pKa = 11.13NPLEE10 pKa = 4.96IDD12 pKa = 3.27TSSYY16 pKa = 11.39VSVCGLPYY24 pKa = 10.61EE25 pKa = 5.4DD26 pKa = 6.06DD27 pKa = 3.99YY28 pKa = 12.05QLSLDD33 pKa = 4.53DD34 pKa = 5.3NGCEE38 pKa = 3.96YY39 pKa = 10.71LKK41 pKa = 10.74KK42 pKa = 10.62VGVIEE47 pKa = 4.38TYY49 pKa = 11.14KK50 pKa = 10.65LIQSHH55 pKa = 6.97RR56 pKa = 11.84DD57 pKa = 3.42SCDD60 pKa = 2.78LAYY63 pKa = 9.8ILDD66 pKa = 4.04TLDD69 pKa = 4.08PDD71 pKa = 4.01SVLGSGSPNVSFDD84 pKa = 4.64DD85 pKa = 4.14IYY87 pKa = 11.05IDD89 pKa = 3.7FTKK92 pKa = 10.63FPTNPGEE99 pKa = 4.13AFNLIKK105 pKa = 10.72NAEE108 pKa = 3.99FAFNKK113 pKa = 10.34LPFEE117 pKa = 4.74LRR119 pKa = 11.84QEE121 pKa = 4.75CNFSPNVFVKK131 pKa = 10.76NLDD134 pKa = 3.35KK135 pKa = 11.04LAEE138 pKa = 4.32KK139 pKa = 10.31FSKK142 pKa = 9.87VDD144 pKa = 3.12KK145 pKa = 10.5VEE147 pKa = 4.2SVGVSNEE154 pKa = 3.56
Molecular weight: 17.4 kDa
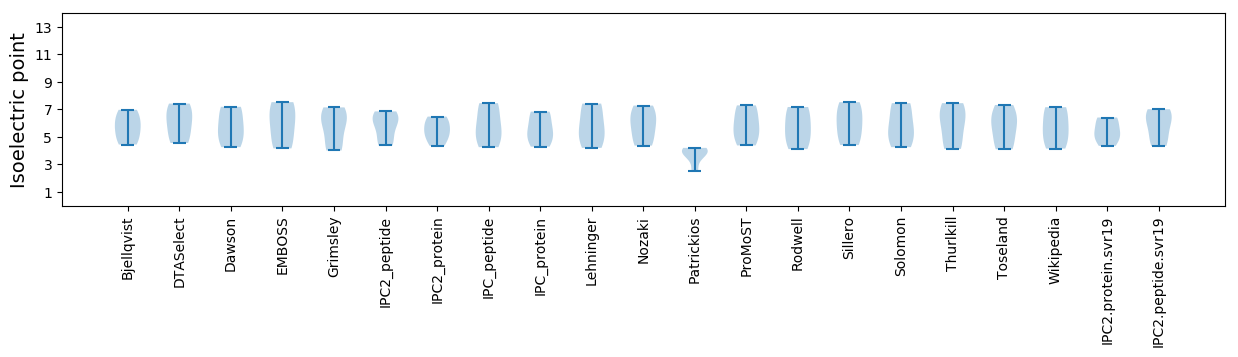
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W543|A0A4P8W543_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_465 OX=2585462 PE=4 SV=1
MM1 pKa = 7.77RR2 pKa = 11.84SILEE6 pKa = 4.2ASQSPNNWFITLTYY20 pKa = 10.94DD21 pKa = 4.06NDD23 pKa = 3.77HH24 pKa = 6.64LPISKK29 pKa = 10.06YY30 pKa = 10.62LNEE33 pKa = 4.04EE34 pKa = 3.86TGVYY38 pKa = 9.83TEE40 pKa = 5.04TPTLDD45 pKa = 3.39NVEE48 pKa = 4.06EE49 pKa = 4.2MKK51 pKa = 10.96KK52 pKa = 8.46FWKK55 pKa = 10.06RR56 pKa = 11.84FRR58 pKa = 11.84INLSRR63 pKa = 11.84SSDD66 pKa = 3.04VDD68 pKa = 3.75LKK70 pKa = 11.28VRR72 pKa = 11.84YY73 pKa = 8.89LYY75 pKa = 10.87SGEE78 pKa = 4.34YY79 pKa = 10.59GDD81 pKa = 3.96TTHH84 pKa = 7.29RR85 pKa = 11.84PHH87 pKa = 6.41FHH89 pKa = 7.26AILFNTVLPDD99 pKa = 3.62LKK101 pKa = 10.99VFYY104 pKa = 10.14KK105 pKa = 10.68DD106 pKa = 2.96KK107 pKa = 10.23QTGFTYY113 pKa = 9.3YY114 pKa = 10.26TSEE117 pKa = 4.37LLEE120 pKa = 4.37KK121 pKa = 10.0SWKK124 pKa = 9.09KK125 pKa = 11.02GNVLIADD132 pKa = 4.27CTFEE136 pKa = 3.95TCQYY140 pKa = 8.4VAAYY144 pKa = 9.24VVKK147 pKa = 10.34KK148 pKa = 7.93QTGKK152 pKa = 10.27QSQVYY157 pKa = 9.9DD158 pKa = 4.17DD159 pKa = 4.39VNIIPPYY166 pKa = 7.81MQMSRR171 pKa = 11.84RR172 pKa = 11.84PGIGSQVIEE181 pKa = 4.58LNLEE185 pKa = 4.4TFWHH189 pKa = 6.87DD190 pKa = 3.17KK191 pKa = 11.03SIILSTKK198 pKa = 10.44NGGKK202 pKa = 9.01QLPFDD207 pKa = 3.68QYY209 pKa = 11.45IKK211 pKa = 11.25NKK213 pKa = 9.85LDD215 pKa = 3.31KK216 pKa = 11.12QFGEE220 pKa = 4.88EE221 pKa = 3.86YY222 pKa = 10.39QEE224 pKa = 4.51YY225 pKa = 10.62KK226 pKa = 10.88NQLCEE231 pKa = 4.67DD232 pKa = 3.6MKK234 pKa = 10.95EE235 pKa = 4.12LQEE238 pKa = 4.1LKK240 pKa = 10.77YY241 pKa = 11.0GSLEE245 pKa = 3.91KK246 pKa = 10.18TYY248 pKa = 10.93LQALLDD254 pKa = 3.74EE255 pKa = 4.79EE256 pKa = 5.71ANFKK260 pKa = 11.14ANYY263 pKa = 8.55FKK265 pKa = 11.25NKK267 pKa = 8.73SRR269 pKa = 11.84KK270 pKa = 8.87LL271 pKa = 3.53
MM1 pKa = 7.77RR2 pKa = 11.84SILEE6 pKa = 4.2ASQSPNNWFITLTYY20 pKa = 10.94DD21 pKa = 4.06NDD23 pKa = 3.77HH24 pKa = 6.64LPISKK29 pKa = 10.06YY30 pKa = 10.62LNEE33 pKa = 4.04EE34 pKa = 3.86TGVYY38 pKa = 9.83TEE40 pKa = 5.04TPTLDD45 pKa = 3.39NVEE48 pKa = 4.06EE49 pKa = 4.2MKK51 pKa = 10.96KK52 pKa = 8.46FWKK55 pKa = 10.06RR56 pKa = 11.84FRR58 pKa = 11.84INLSRR63 pKa = 11.84SSDD66 pKa = 3.04VDD68 pKa = 3.75LKK70 pKa = 11.28VRR72 pKa = 11.84YY73 pKa = 8.89LYY75 pKa = 10.87SGEE78 pKa = 4.34YY79 pKa = 10.59GDD81 pKa = 3.96TTHH84 pKa = 7.29RR85 pKa = 11.84PHH87 pKa = 6.41FHH89 pKa = 7.26AILFNTVLPDD99 pKa = 3.62LKK101 pKa = 10.99VFYY104 pKa = 10.14KK105 pKa = 10.68DD106 pKa = 2.96KK107 pKa = 10.23QTGFTYY113 pKa = 9.3YY114 pKa = 10.26TSEE117 pKa = 4.37LLEE120 pKa = 4.37KK121 pKa = 10.0SWKK124 pKa = 9.09KK125 pKa = 11.02GNVLIADD132 pKa = 4.27CTFEE136 pKa = 3.95TCQYY140 pKa = 8.4VAAYY144 pKa = 9.24VVKK147 pKa = 10.34KK148 pKa = 7.93QTGKK152 pKa = 10.27QSQVYY157 pKa = 9.9DD158 pKa = 4.17DD159 pKa = 4.39VNIIPPYY166 pKa = 7.81MQMSRR171 pKa = 11.84RR172 pKa = 11.84PGIGSQVIEE181 pKa = 4.58LNLEE185 pKa = 4.4TFWHH189 pKa = 6.87DD190 pKa = 3.17KK191 pKa = 11.03SIILSTKK198 pKa = 10.44NGGKK202 pKa = 9.01QLPFDD207 pKa = 3.68QYY209 pKa = 11.45IKK211 pKa = 11.25NKK213 pKa = 9.85LDD215 pKa = 3.31KK216 pKa = 11.12QFGEE220 pKa = 4.88EE221 pKa = 3.86YY222 pKa = 10.39QEE224 pKa = 4.51YY225 pKa = 10.62KK226 pKa = 10.88NQLCEE231 pKa = 4.67DD232 pKa = 3.6MKK234 pKa = 10.95EE235 pKa = 4.12LQEE238 pKa = 4.1LKK240 pKa = 10.77YY241 pKa = 11.0GSLEE245 pKa = 3.91KK246 pKa = 10.18TYY248 pKa = 10.93LQALLDD254 pKa = 3.74EE255 pKa = 4.79EE256 pKa = 5.71ANFKK260 pKa = 11.14ANYY263 pKa = 8.55FKK265 pKa = 11.25NKK267 pKa = 8.73SRR269 pKa = 11.84KK270 pKa = 8.87LL271 pKa = 3.53
Molecular weight: 31.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1463 |
82 |
561 |
243.8 |
27.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.177 ± 2.799 | 1.025 ± 0.296 |
6.083 ± 0.837 | 4.99 ± 1.007 |
4.785 ± 0.849 | 6.015 ± 0.77 |
1.846 ± 0.589 | 4.785 ± 0.387 |
5.947 ± 1.433 | 8.271 ± 0.895 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.392 ± 0.329 | 6.63 ± 1.038 |
3.623 ± 0.865 | 4.238 ± 0.741 |
3.008 ± 0.524 | 10.321 ± 1.578 |
6.015 ± 0.707 | 6.425 ± 0.675 |
1.025 ± 0.423 | 5.4 ± 0.554 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
