
Zymoseptoria tritici ST99CH_3D7
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetidae; Mycosphaerellales; Mycosphaerellaceae; Zymoseptoria; Zymoseptoria tritici
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
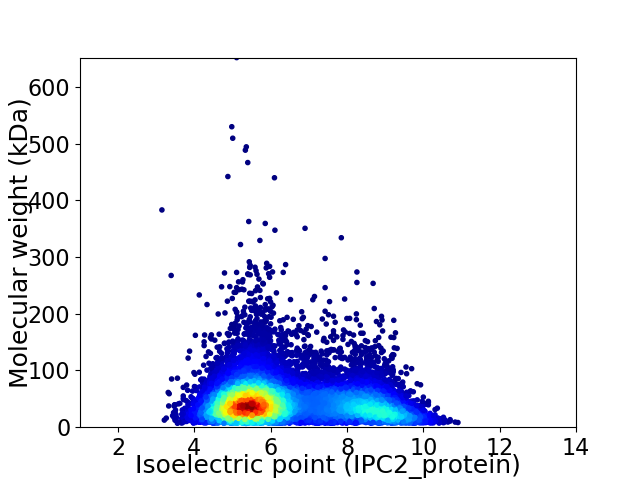
Virtual 2D-PAGE plot for 11722 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X7RT76|A0A1X7RT76_ZYMTR AA_permease domain-containing protein OS=Zymoseptoria tritici ST99CH_3D7 OX=1276538 GN=ZT3D7_G5785 PE=4 SV=1
MM1 pKa = 7.36GLAASSVSFEE11 pKa = 4.17SMDD14 pKa = 3.12VGGAVLLQDD23 pKa = 3.27GVVPVFLTSYY33 pKa = 10.39GFSPANMLSFGVIAALSYY51 pKa = 9.33FSPAFFFGVTDD62 pKa = 3.25TDD64 pKa = 4.37YY65 pKa = 11.56IDD67 pKa = 3.76VQPVLTFPVIDD78 pKa = 3.99AGGAASTSVVTCAPFNNGMLPPPEE102 pKa = 4.29SSGVDD107 pKa = 3.03PVVYY111 pKa = 9.27ITATNQLKK119 pKa = 10.62CSIPNTSIAPGVVTIAFEE137 pKa = 4.12KK138 pKa = 10.58SDD140 pKa = 4.13GIFFTDD146 pKa = 3.34ASYY149 pKa = 11.57GFLVADD155 pKa = 3.72STILADD161 pKa = 3.86TVTATNTVFVPGSTVGSTTVTSTRR185 pKa = 11.84YY186 pKa = 9.89IFTDD190 pKa = 3.27EE191 pKa = 4.19ATSTSTTTLSDD202 pKa = 3.39ASSTTYY208 pKa = 10.48ISCSTSTSATNTGTATADD226 pKa = 3.72PNSTSATDD234 pKa = 3.42SSSITSATDD243 pKa = 3.72LSSTTSATDD252 pKa = 3.13SSNTSSSATDD262 pKa = 3.56DD263 pKa = 3.63SSSITISDD271 pKa = 3.78TPTAPTYY278 pKa = 8.26PTDD281 pKa = 3.84SNPFSSSTSCTNPQTPSPTPQKK303 pKa = 10.48NIKK306 pKa = 8.62TVFITDD312 pKa = 3.5CAKK315 pKa = 10.51AAKK318 pKa = 9.25TSAAAAALSARR329 pKa = 11.84QTTATDD335 pKa = 3.44TLASNFGAPDD345 pKa = 3.56FTFSNGDD352 pKa = 3.07NTATITVTSTAIDD365 pKa = 3.49TRR367 pKa = 11.84STVVTSGTDD376 pKa = 3.17TSVVTEE382 pKa = 4.31SDD384 pKa = 3.26PTTTRR389 pKa = 11.84TVTAATVDD397 pKa = 3.42EE398 pKa = 4.55RR399 pKa = 11.84TVEE402 pKa = 4.23TVVGCEE408 pKa = 3.81GSEE411 pKa = 4.08TARR414 pKa = 3.84
MM1 pKa = 7.36GLAASSVSFEE11 pKa = 4.17SMDD14 pKa = 3.12VGGAVLLQDD23 pKa = 3.27GVVPVFLTSYY33 pKa = 10.39GFSPANMLSFGVIAALSYY51 pKa = 9.33FSPAFFFGVTDD62 pKa = 3.25TDD64 pKa = 4.37YY65 pKa = 11.56IDD67 pKa = 3.76VQPVLTFPVIDD78 pKa = 3.99AGGAASTSVVTCAPFNNGMLPPPEE102 pKa = 4.29SSGVDD107 pKa = 3.03PVVYY111 pKa = 9.27ITATNQLKK119 pKa = 10.62CSIPNTSIAPGVVTIAFEE137 pKa = 4.12KK138 pKa = 10.58SDD140 pKa = 4.13GIFFTDD146 pKa = 3.34ASYY149 pKa = 11.57GFLVADD155 pKa = 3.72STILADD161 pKa = 3.86TVTATNTVFVPGSTVGSTTVTSTRR185 pKa = 11.84YY186 pKa = 9.89IFTDD190 pKa = 3.27EE191 pKa = 4.19ATSTSTTTLSDD202 pKa = 3.39ASSTTYY208 pKa = 10.48ISCSTSTSATNTGTATADD226 pKa = 3.72PNSTSATDD234 pKa = 3.42SSSITSATDD243 pKa = 3.72LSSTTSATDD252 pKa = 3.13SSNTSSSATDD262 pKa = 3.56DD263 pKa = 3.63SSSITISDD271 pKa = 3.78TPTAPTYY278 pKa = 8.26PTDD281 pKa = 3.84SNPFSSSTSCTNPQTPSPTPQKK303 pKa = 10.48NIKK306 pKa = 8.62TVFITDD312 pKa = 3.5CAKK315 pKa = 10.51AAKK318 pKa = 9.25TSAAAAALSARR329 pKa = 11.84QTTATDD335 pKa = 3.44TLASNFGAPDD345 pKa = 3.56FTFSNGDD352 pKa = 3.07NTATITVTSTAIDD365 pKa = 3.49TRR367 pKa = 11.84STVVTSGTDD376 pKa = 3.17TSVVTEE382 pKa = 4.31SDD384 pKa = 3.26PTTTRR389 pKa = 11.84TVTAATVDD397 pKa = 3.42EE398 pKa = 4.55RR399 pKa = 11.84TVEE402 pKa = 4.23TVVGCEE408 pKa = 3.81GSEE411 pKa = 4.08TARR414 pKa = 3.84
Molecular weight: 41.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X7RQL8|A0A1X7RQL8_ZYMTR NADH-cytochrome b5 reductase OS=Zymoseptoria tritici ST99CH_3D7 OX=1276538 GN=ZT3D7_G4895 PE=3 SV=1
MM1 pKa = 7.59SYY3 pKa = 10.56LARR6 pKa = 11.84RR7 pKa = 11.84AAATVRR13 pKa = 11.84LANTITTRR21 pKa = 11.84RR22 pKa = 11.84GLVGISAGRR31 pKa = 11.84AATIVRR37 pKa = 11.84LTSNAPGRR45 pKa = 11.84IHH47 pKa = 6.96ARR49 pKa = 11.84STARR53 pKa = 11.84RR54 pKa = 11.84AVPGVRR60 pKa = 11.84LVNTAWRR67 pKa = 11.84PDD69 pKa = 3.3MPAQGSSIAA78 pKa = 3.92
MM1 pKa = 7.59SYY3 pKa = 10.56LARR6 pKa = 11.84RR7 pKa = 11.84AAATVRR13 pKa = 11.84LANTITTRR21 pKa = 11.84RR22 pKa = 11.84GLVGISAGRR31 pKa = 11.84AATIVRR37 pKa = 11.84LTSNAPGRR45 pKa = 11.84IHH47 pKa = 6.96ARR49 pKa = 11.84STARR53 pKa = 11.84RR54 pKa = 11.84AVPGVRR60 pKa = 11.84LVNTAWRR67 pKa = 11.84PDD69 pKa = 3.3MPAQGSSIAA78 pKa = 3.92
Molecular weight: 8.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5360897 |
66 |
6000 |
457.3 |
50.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.154 ± 0.021 | 1.21 ± 0.009 |
5.847 ± 0.019 | 6.347 ± 0.024 |
3.532 ± 0.014 | 7.047 ± 0.023 |
2.396 ± 0.01 | 4.533 ± 0.013 |
4.903 ± 0.021 | 8.565 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.236 ± 0.009 | 3.589 ± 0.012 |
6.029 ± 0.024 | 4.101 ± 0.017 |
6.169 ± 0.023 | 8.117 ± 0.029 |
6.154 ± 0.022 | 6.036 ± 0.014 |
1.412 ± 0.009 | 2.622 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
