
Allobacillus sp. SKP2-8
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Allobacillus; unclassified Allobacillus
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins
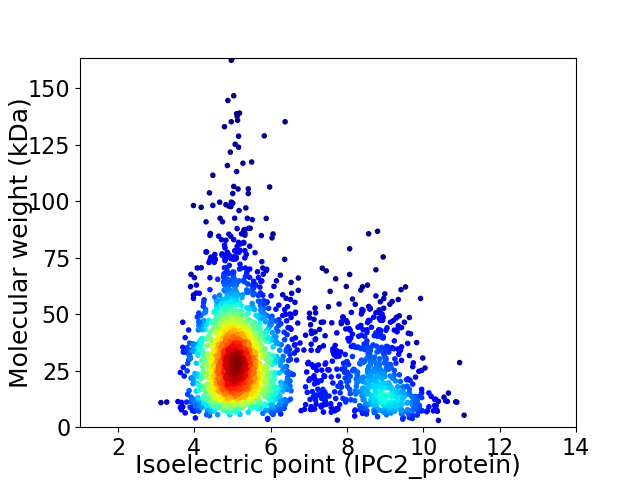
Virtual 2D-PAGE plot for 2475 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A556PNV1|A0A556PNV1_9BACI 2-keto-3-deoxygluconate permease OS=Allobacillus sp. SKP2-8 OX=1955271 GN=FPQ10_07940 PE=3 SV=1
MM1 pKa = 7.8LFFLSLALLLVIAACGGSDD20 pKa = 3.77EE21 pKa = 5.21DD22 pKa = 4.37SSSGSEE28 pKa = 3.93GEE30 pKa = 4.28EE31 pKa = 3.82NGEE34 pKa = 4.16EE35 pKa = 4.16SSSEE39 pKa = 4.0SSSEE43 pKa = 3.94DD44 pKa = 3.56GEE46 pKa = 4.29SSEE49 pKa = 5.32ASGEE53 pKa = 4.27SYY55 pKa = 10.67EE56 pKa = 5.56LEE58 pKa = 4.66FGMQPNTQANEE69 pKa = 3.72YY70 pKa = 9.73KK71 pKa = 10.54AAEE74 pKa = 4.0FLADD78 pKa = 3.84YY79 pKa = 10.75VEE81 pKa = 4.62KK82 pKa = 10.82EE83 pKa = 3.59SDD85 pKa = 3.21GRR87 pKa = 11.84LTVTIFPDD95 pKa = 3.4AQLGNDD101 pKa = 3.72LSMLGQLQDD110 pKa = 3.33GTLDD114 pKa = 3.11ITLAEE119 pKa = 4.11MGRR122 pKa = 11.84FGEE125 pKa = 4.39WIPRR129 pKa = 11.84AEE131 pKa = 4.58LLAMPYY137 pKa = 10.2VIEE140 pKa = 5.34DD141 pKa = 3.25FDD143 pKa = 4.56HH144 pKa = 6.66IKK146 pKa = 10.78NVVYY150 pKa = 8.91NTEE153 pKa = 3.86FGEE156 pKa = 4.16EE157 pKa = 3.65LRR159 pKa = 11.84AEE161 pKa = 4.18LVEE164 pKa = 4.01EE165 pKa = 3.97HH166 pKa = 6.3GMRR169 pKa = 11.84VIDD172 pKa = 3.68SAYY175 pKa = 9.61NGPRR179 pKa = 11.84VTSSNSPIEE188 pKa = 4.13EE189 pKa = 4.13LADD192 pKa = 3.59MEE194 pKa = 4.5GMSLRR199 pKa = 11.84VPEE202 pKa = 4.53AQTLLDD208 pKa = 3.57YY209 pKa = 11.59AEE211 pKa = 4.14YY212 pKa = 9.94TGANPTPMAFGEE224 pKa = 4.67VYY226 pKa = 10.6LALQTGQVDD235 pKa = 4.05GQEE238 pKa = 4.28NPLPTIEE245 pKa = 4.28AQSFHH250 pKa = 6.53EE251 pKa = 4.23VQDD254 pKa = 4.21YY255 pKa = 10.78IALTNHH261 pKa = 5.12VVNDD265 pKa = 3.33NTYY268 pKa = 8.8VVSEE272 pKa = 4.4KK273 pKa = 10.12TYY275 pKa = 10.81QDD277 pKa = 3.44LPEE280 pKa = 4.63DD281 pKa = 3.39LRR283 pKa = 11.84TILEE287 pKa = 4.12EE288 pKa = 5.18GIDD291 pKa = 3.7AATDD295 pKa = 3.05HH296 pKa = 6.24HH297 pKa = 6.9TEE299 pKa = 4.2LFQTQEE305 pKa = 4.08DD306 pKa = 4.19EE307 pKa = 4.69LISFFEE313 pKa = 4.33EE314 pKa = 3.71EE315 pKa = 4.79GVTITEE321 pKa = 4.21PNLDD325 pKa = 3.82EE326 pKa = 4.47FRR328 pKa = 11.84EE329 pKa = 4.63AVSEE333 pKa = 4.36AYY335 pKa = 9.39PSYY338 pKa = 11.16YY339 pKa = 10.45EE340 pKa = 5.98DD341 pKa = 3.2IGEE344 pKa = 4.24DD345 pKa = 3.16AEE347 pKa = 5.15KK348 pKa = 10.76YY349 pKa = 9.51MEE351 pKa = 4.7VIQGAAEE358 pKa = 3.79
MM1 pKa = 7.8LFFLSLALLLVIAACGGSDD20 pKa = 3.77EE21 pKa = 5.21DD22 pKa = 4.37SSSGSEE28 pKa = 3.93GEE30 pKa = 4.28EE31 pKa = 3.82NGEE34 pKa = 4.16EE35 pKa = 4.16SSSEE39 pKa = 4.0SSSEE43 pKa = 3.94DD44 pKa = 3.56GEE46 pKa = 4.29SSEE49 pKa = 5.32ASGEE53 pKa = 4.27SYY55 pKa = 10.67EE56 pKa = 5.56LEE58 pKa = 4.66FGMQPNTQANEE69 pKa = 3.72YY70 pKa = 9.73KK71 pKa = 10.54AAEE74 pKa = 4.0FLADD78 pKa = 3.84YY79 pKa = 10.75VEE81 pKa = 4.62KK82 pKa = 10.82EE83 pKa = 3.59SDD85 pKa = 3.21GRR87 pKa = 11.84LTVTIFPDD95 pKa = 3.4AQLGNDD101 pKa = 3.72LSMLGQLQDD110 pKa = 3.33GTLDD114 pKa = 3.11ITLAEE119 pKa = 4.11MGRR122 pKa = 11.84FGEE125 pKa = 4.39WIPRR129 pKa = 11.84AEE131 pKa = 4.58LLAMPYY137 pKa = 10.2VIEE140 pKa = 5.34DD141 pKa = 3.25FDD143 pKa = 4.56HH144 pKa = 6.66IKK146 pKa = 10.78NVVYY150 pKa = 8.91NTEE153 pKa = 3.86FGEE156 pKa = 4.16EE157 pKa = 3.65LRR159 pKa = 11.84AEE161 pKa = 4.18LVEE164 pKa = 4.01EE165 pKa = 3.97HH166 pKa = 6.3GMRR169 pKa = 11.84VIDD172 pKa = 3.68SAYY175 pKa = 9.61NGPRR179 pKa = 11.84VTSSNSPIEE188 pKa = 4.13EE189 pKa = 4.13LADD192 pKa = 3.59MEE194 pKa = 4.5GMSLRR199 pKa = 11.84VPEE202 pKa = 4.53AQTLLDD208 pKa = 3.57YY209 pKa = 11.59AEE211 pKa = 4.14YY212 pKa = 9.94TGANPTPMAFGEE224 pKa = 4.67VYY226 pKa = 10.6LALQTGQVDD235 pKa = 4.05GQEE238 pKa = 4.28NPLPTIEE245 pKa = 4.28AQSFHH250 pKa = 6.53EE251 pKa = 4.23VQDD254 pKa = 4.21YY255 pKa = 10.78IALTNHH261 pKa = 5.12VVNDD265 pKa = 3.33NTYY268 pKa = 8.8VVSEE272 pKa = 4.4KK273 pKa = 10.12TYY275 pKa = 10.81QDD277 pKa = 3.44LPEE280 pKa = 4.63DD281 pKa = 3.39LRR283 pKa = 11.84TILEE287 pKa = 4.12EE288 pKa = 5.18GIDD291 pKa = 3.7AATDD295 pKa = 3.05HH296 pKa = 6.24HH297 pKa = 6.9TEE299 pKa = 4.2LFQTQEE305 pKa = 4.08DD306 pKa = 4.19EE307 pKa = 4.69LISFFEE313 pKa = 4.33EE314 pKa = 3.71EE315 pKa = 4.79GVTITEE321 pKa = 4.21PNLDD325 pKa = 3.82EE326 pKa = 4.47FRR328 pKa = 11.84EE329 pKa = 4.63AVSEE333 pKa = 4.36AYY335 pKa = 9.39PSYY338 pKa = 11.16YY339 pKa = 10.45EE340 pKa = 5.98DD341 pKa = 3.2IGEE344 pKa = 4.24DD345 pKa = 3.16AEE347 pKa = 5.15KK348 pKa = 10.76YY349 pKa = 9.51MEE351 pKa = 4.7VIQGAAEE358 pKa = 3.79
Molecular weight: 39.65 kDa
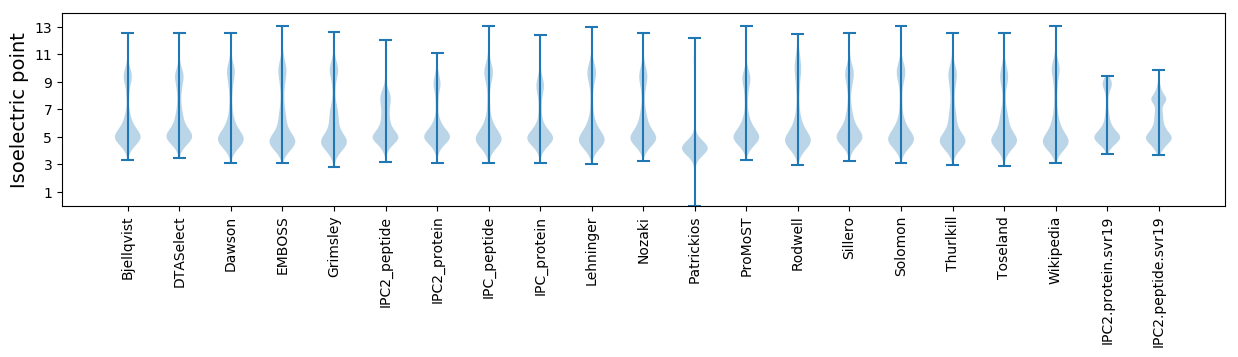
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A556PMV2|A0A556PMV2_9BACI TIGR02678 family protein OS=Allobacillus sp. SKP2-8 OX=1955271 GN=FPQ10_08795 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.54QPKK8 pKa = 9.44KK9 pKa = 7.96RR10 pKa = 11.84KK11 pKa = 8.69HH12 pKa = 5.94SKK14 pKa = 8.54VHH16 pKa = 5.76GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.78TKK25 pKa = 10.39NGRR28 pKa = 11.84KK29 pKa = 8.66VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.2KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.54QPKK8 pKa = 9.44KK9 pKa = 7.96RR10 pKa = 11.84KK11 pKa = 8.69HH12 pKa = 5.94SKK14 pKa = 8.54VHH16 pKa = 5.76GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 8.78TKK25 pKa = 10.39NGRR28 pKa = 11.84KK29 pKa = 8.66VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84KK36 pKa = 9.2KK37 pKa = 10.05GRR39 pKa = 11.84KK40 pKa = 8.66VLSAA44 pKa = 4.05
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
700000 |
25 |
1429 |
282.8 |
31.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.523 ± 0.047 | 0.562 ± 0.014 |
5.59 ± 0.042 | 8.287 ± 0.062 |
4.605 ± 0.043 | 6.639 ± 0.047 |
2.129 ± 0.024 | 7.712 ± 0.049 |
6.461 ± 0.048 | 9.538 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.848 ± 0.026 | 4.398 ± 0.033 |
3.525 ± 0.031 | 4.221 ± 0.043 |
4.191 ± 0.035 | 5.89 ± 0.035 |
5.401 ± 0.033 | 7.021 ± 0.04 |
0.961 ± 0.017 | 3.498 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
