
Wuhan insect virus 35
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.63
Get precalculated fractions of proteins
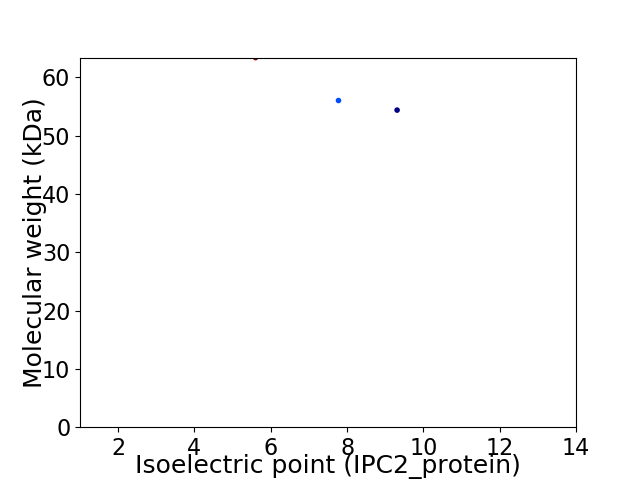
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGP0|A0A1L3KGP0_9VIRU DiSB-ORF2_chro domain-containing protein OS=Wuhan insect virus 35 OX=1923738 PE=4 SV=1
MM1 pKa = 8.3DD2 pKa = 4.35YY3 pKa = 10.97LRR5 pKa = 11.84QLMQNFIGGEE15 pKa = 3.86PDD17 pKa = 3.35EE18 pKa = 5.35LPEE21 pKa = 4.08EE22 pKa = 4.17QAVEE26 pKa = 4.45RR27 pKa = 11.84EE28 pKa = 4.2TQLDD32 pKa = 3.76QIINTLDD39 pKa = 3.14RR40 pKa = 11.84LRR42 pKa = 11.84RR43 pKa = 11.84VQQLTHH49 pKa = 6.9EE50 pKa = 4.52IDD52 pKa = 3.95DD53 pKa = 4.29ARR55 pKa = 11.84TAPIPQGEE63 pKa = 4.33TAEE66 pKa = 4.06EE67 pKa = 3.93RR68 pKa = 11.84VLRR71 pKa = 11.84GVRR74 pKa = 11.84AQAEE78 pKa = 4.76VIEE81 pKa = 4.26HH82 pKa = 5.94RR83 pKa = 11.84QVIDD87 pKa = 4.68DD88 pKa = 3.8KK89 pKa = 9.5TTPPSVDD96 pKa = 2.93DD97 pKa = 4.65CYY99 pKa = 11.85AFLKK103 pKa = 9.0TRR105 pKa = 11.84CEE107 pKa = 4.01GKK109 pKa = 10.63LVDD112 pKa = 3.9GTYY115 pKa = 10.69NAYY118 pKa = 9.93CAALIRR124 pKa = 11.84QYY126 pKa = 10.9FATRR130 pKa = 11.84KK131 pKa = 8.39PVSNKK136 pKa = 8.8MEE138 pKa = 4.08YY139 pKa = 10.69AIMNSVMNMHH149 pKa = 7.87LINIRR154 pKa = 11.84PQYY157 pKa = 11.02DD158 pKa = 3.62YY159 pKa = 11.3DD160 pKa = 3.53LHH162 pKa = 7.74DD163 pKa = 3.59ILRR166 pKa = 11.84YY167 pKa = 9.33NDD169 pKa = 4.15CINYY173 pKa = 7.71VQRR176 pKa = 11.84KK177 pKa = 5.69TIRR180 pKa = 11.84IFNQIIEE187 pKa = 4.26IPFTRR192 pKa = 11.84ITLEE196 pKa = 3.97DD197 pKa = 3.85LNIFRR202 pKa = 11.84PAATSRR208 pKa = 11.84PINFLWKK215 pKa = 10.48LFLALSLIIIAYY227 pKa = 7.95YY228 pKa = 9.98AVHH231 pKa = 7.42FITLPFTLLTTFTTTHH247 pKa = 6.15NQPQQPSQPIITATDD262 pKa = 3.41PSYY265 pKa = 11.14YY266 pKa = 9.04QTITHH271 pKa = 7.68LISTWTTSSTSSNDD285 pKa = 3.72DD286 pKa = 3.51YY287 pKa = 12.11SHH289 pKa = 6.94GPLSSIYY296 pKa = 9.36HH297 pKa = 4.74QWIAPASEE305 pKa = 4.32SFTRR309 pKa = 11.84TLTLVYY315 pKa = 10.8YY316 pKa = 10.63KK317 pKa = 10.22DD318 pKa = 3.62AKK320 pKa = 10.0PALEE324 pKa = 4.3SLLSLSWRR332 pKa = 11.84KK333 pKa = 8.65CQHH336 pKa = 6.65LNTKK340 pKa = 10.08LQDD343 pKa = 3.57LYY345 pKa = 11.63KK346 pKa = 10.63EE347 pKa = 4.45GTPHH351 pKa = 6.53LTYY354 pKa = 11.47AMDD357 pKa = 5.76AISNHH362 pKa = 4.82WNAVSKK368 pKa = 10.21ATDD371 pKa = 3.35TLAKK375 pKa = 10.09EE376 pKa = 4.18LTTILAAKK384 pKa = 7.4YY385 pKa = 8.15TSSLANINTTLRR397 pKa = 11.84EE398 pKa = 4.12TILPLTLTSLSKK410 pKa = 10.97CYY412 pKa = 10.49DD413 pKa = 3.39YY414 pKa = 11.06ATDD417 pKa = 4.62FIYY420 pKa = 10.74DD421 pKa = 3.78VTNKK425 pKa = 10.08APNYY429 pKa = 9.04LSSVKK434 pKa = 10.25KK435 pKa = 10.33QLEE438 pKa = 4.52TEE440 pKa = 4.62LLPAMEE446 pKa = 5.44KK447 pKa = 10.29SIQYY451 pKa = 9.65AAQEE455 pKa = 4.46CPAMWTQALEE465 pKa = 4.07IASSIITSSGMLLNDD480 pKa = 5.19FISTVMRR487 pKa = 11.84LSTEE491 pKa = 4.36TISSYY496 pKa = 11.24SQTLKK501 pKa = 10.73SQQQNSQTYY510 pKa = 8.47YY511 pKa = 11.19GSTIWKK517 pKa = 7.89QLLVIVLILYY527 pKa = 7.62TKK529 pKa = 10.91LNFAVADD536 pKa = 3.54WFIIPTAIPQWPLTRR551 pKa = 11.84IDD553 pKa = 3.22
MM1 pKa = 8.3DD2 pKa = 4.35YY3 pKa = 10.97LRR5 pKa = 11.84QLMQNFIGGEE15 pKa = 3.86PDD17 pKa = 3.35EE18 pKa = 5.35LPEE21 pKa = 4.08EE22 pKa = 4.17QAVEE26 pKa = 4.45RR27 pKa = 11.84EE28 pKa = 4.2TQLDD32 pKa = 3.76QIINTLDD39 pKa = 3.14RR40 pKa = 11.84LRR42 pKa = 11.84RR43 pKa = 11.84VQQLTHH49 pKa = 6.9EE50 pKa = 4.52IDD52 pKa = 3.95DD53 pKa = 4.29ARR55 pKa = 11.84TAPIPQGEE63 pKa = 4.33TAEE66 pKa = 4.06EE67 pKa = 3.93RR68 pKa = 11.84VLRR71 pKa = 11.84GVRR74 pKa = 11.84AQAEE78 pKa = 4.76VIEE81 pKa = 4.26HH82 pKa = 5.94RR83 pKa = 11.84QVIDD87 pKa = 4.68DD88 pKa = 3.8KK89 pKa = 9.5TTPPSVDD96 pKa = 2.93DD97 pKa = 4.65CYY99 pKa = 11.85AFLKK103 pKa = 9.0TRR105 pKa = 11.84CEE107 pKa = 4.01GKK109 pKa = 10.63LVDD112 pKa = 3.9GTYY115 pKa = 10.69NAYY118 pKa = 9.93CAALIRR124 pKa = 11.84QYY126 pKa = 10.9FATRR130 pKa = 11.84KK131 pKa = 8.39PVSNKK136 pKa = 8.8MEE138 pKa = 4.08YY139 pKa = 10.69AIMNSVMNMHH149 pKa = 7.87LINIRR154 pKa = 11.84PQYY157 pKa = 11.02DD158 pKa = 3.62YY159 pKa = 11.3DD160 pKa = 3.53LHH162 pKa = 7.74DD163 pKa = 3.59ILRR166 pKa = 11.84YY167 pKa = 9.33NDD169 pKa = 4.15CINYY173 pKa = 7.71VQRR176 pKa = 11.84KK177 pKa = 5.69TIRR180 pKa = 11.84IFNQIIEE187 pKa = 4.26IPFTRR192 pKa = 11.84ITLEE196 pKa = 3.97DD197 pKa = 3.85LNIFRR202 pKa = 11.84PAATSRR208 pKa = 11.84PINFLWKK215 pKa = 10.48LFLALSLIIIAYY227 pKa = 7.95YY228 pKa = 9.98AVHH231 pKa = 7.42FITLPFTLLTTFTTTHH247 pKa = 6.15NQPQQPSQPIITATDD262 pKa = 3.41PSYY265 pKa = 11.14YY266 pKa = 9.04QTITHH271 pKa = 7.68LISTWTTSSTSSNDD285 pKa = 3.72DD286 pKa = 3.51YY287 pKa = 12.11SHH289 pKa = 6.94GPLSSIYY296 pKa = 9.36HH297 pKa = 4.74QWIAPASEE305 pKa = 4.32SFTRR309 pKa = 11.84TLTLVYY315 pKa = 10.8YY316 pKa = 10.63KK317 pKa = 10.22DD318 pKa = 3.62AKK320 pKa = 10.0PALEE324 pKa = 4.3SLLSLSWRR332 pKa = 11.84KK333 pKa = 8.65CQHH336 pKa = 6.65LNTKK340 pKa = 10.08LQDD343 pKa = 3.57LYY345 pKa = 11.63KK346 pKa = 10.63EE347 pKa = 4.45GTPHH351 pKa = 6.53LTYY354 pKa = 11.47AMDD357 pKa = 5.76AISNHH362 pKa = 4.82WNAVSKK368 pKa = 10.21ATDD371 pKa = 3.35TLAKK375 pKa = 10.09EE376 pKa = 4.18LTTILAAKK384 pKa = 7.4YY385 pKa = 8.15TSSLANINTTLRR397 pKa = 11.84EE398 pKa = 4.12TILPLTLTSLSKK410 pKa = 10.97CYY412 pKa = 10.49DD413 pKa = 3.39YY414 pKa = 11.06ATDD417 pKa = 4.62FIYY420 pKa = 10.74DD421 pKa = 3.78VTNKK425 pKa = 10.08APNYY429 pKa = 9.04LSSVKK434 pKa = 10.25KK435 pKa = 10.33QLEE438 pKa = 4.52TEE440 pKa = 4.62LLPAMEE446 pKa = 5.44KK447 pKa = 10.29SIQYY451 pKa = 9.65AAQEE455 pKa = 4.46CPAMWTQALEE465 pKa = 4.07IASSIITSSGMLLNDD480 pKa = 5.19FISTVMRR487 pKa = 11.84LSTEE491 pKa = 4.36TISSYY496 pKa = 11.24SQTLKK501 pKa = 10.73SQQQNSQTYY510 pKa = 8.47YY511 pKa = 11.19GSTIWKK517 pKa = 7.89QLLVIVLILYY527 pKa = 7.62TKK529 pKa = 10.91LNFAVADD536 pKa = 3.54WFIIPTAIPQWPLTRR551 pKa = 11.84IDD553 pKa = 3.22
Molecular weight: 63.36 kDa
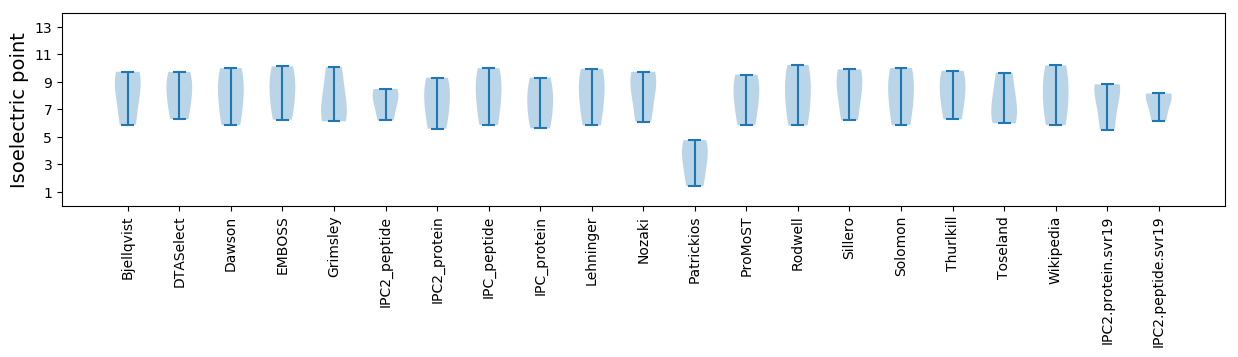
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH03|A0A1L3KH03_9VIRU RNA-directed RNA polymerase OS=Wuhan insect virus 35 OX=1923738 PE=4 SV=1
MM1 pKa = 6.87EE2 pKa = 4.95TVPGIITNHH11 pKa = 4.84NRR13 pKa = 11.84LLRR16 pKa = 11.84RR17 pKa = 11.84TLYY20 pKa = 10.36HH21 pKa = 6.13PPIYY25 pKa = 10.3APHH28 pKa = 7.12DD29 pKa = 3.95FYY31 pKa = 11.79YY32 pKa = 10.17YY33 pKa = 10.46AQSATTTIAAYY44 pKa = 10.07HH45 pKa = 5.67NRR47 pKa = 11.84HH48 pKa = 5.94RR49 pKa = 11.84PIILPDD55 pKa = 3.48YY56 pKa = 10.25NPSHH60 pKa = 6.91FNVDD64 pKa = 3.53NIFDD68 pKa = 3.69KK69 pKa = 10.97FKK71 pKa = 11.02RR72 pKa = 11.84RR73 pKa = 11.84LQPWTFEE80 pKa = 4.14QYY82 pKa = 10.08ISSMDD87 pKa = 3.45SPRR90 pKa = 11.84KK91 pKa = 9.33RR92 pKa = 11.84KK93 pKa = 9.4FYY95 pKa = 10.56QDD97 pKa = 3.59AYY99 pKa = 9.98TSILQGRR106 pKa = 11.84KK107 pKa = 6.77TSSGITPFTKK117 pKa = 10.25LEE119 pKa = 3.97KK120 pKa = 10.05MSTSKK125 pKa = 11.12YY126 pKa = 9.02KK127 pKa = 10.62APRR130 pKa = 11.84LIQGRR135 pKa = 11.84HH136 pKa = 3.97PTFNLCYY143 pKa = 10.14GRR145 pKa = 11.84YY146 pKa = 8.84IKK148 pKa = 10.4PLEE151 pKa = 4.32RR152 pKa = 11.84SLKK155 pKa = 9.73GDD157 pKa = 3.28RR158 pKa = 11.84HH159 pKa = 5.76FGKK162 pKa = 9.03GTYY165 pKa = 10.34DD166 pKa = 3.93DD167 pKa = 4.1IGRR170 pKa = 11.84KK171 pKa = 7.15IHH173 pKa = 6.35KK174 pKa = 10.12LSRR177 pKa = 11.84KK178 pKa = 7.88YY179 pKa = 10.51KK180 pKa = 8.49YY181 pKa = 9.07YY182 pKa = 10.7TEE184 pKa = 5.02GDD186 pKa = 3.3HH187 pKa = 5.74TTFDD191 pKa = 3.49SHH193 pKa = 5.5VTVEE197 pKa = 4.04MLRR200 pKa = 11.84LCHH203 pKa = 6.7RR204 pKa = 11.84FYY206 pKa = 11.23LRR208 pKa = 11.84CYY210 pKa = 8.62KK211 pKa = 10.29QSPEE215 pKa = 4.27LLKK218 pKa = 10.88LCKK221 pKa = 9.35KK222 pKa = 8.68TIRR225 pKa = 11.84NRR227 pKa = 11.84AVARR231 pKa = 11.84NGEE234 pKa = 4.39KK235 pKa = 9.42YY236 pKa = 7.83TVRR239 pKa = 11.84GTRR242 pKa = 11.84MSGDD246 pKa = 3.12VDD248 pKa = 3.51TGFGNSIINYY258 pKa = 8.11YY259 pKa = 9.81IIRR262 pKa = 11.84HH263 pKa = 4.72ALKK266 pKa = 10.19RR267 pKa = 11.84LHH269 pKa = 6.9IDD271 pKa = 2.92GDD273 pKa = 4.42AIVNGDD279 pKa = 3.89DD280 pKa = 5.52FIIFTNIKK288 pKa = 9.62ISTAEE293 pKa = 3.74FTNLLRR299 pKa = 11.84EE300 pKa = 4.34YY301 pKa = 11.41NMEE304 pKa = 4.07TTLGNSVDD312 pKa = 4.58SIHH315 pKa = 6.22QVEE318 pKa = 5.49FCRR321 pKa = 11.84CRR323 pKa = 11.84LVYY326 pKa = 10.5HH327 pKa = 7.16PDD329 pKa = 3.02GHH331 pKa = 6.34PTMAFDD337 pKa = 3.46PHH339 pKa = 8.13RR340 pKa = 11.84LEE342 pKa = 5.61KK343 pKa = 10.19IYY345 pKa = 10.86GCSNILYY352 pKa = 7.75PKK354 pKa = 8.93KK355 pKa = 10.35KK356 pKa = 10.21KK357 pKa = 10.82YY358 pKa = 10.31EE359 pKa = 4.49DD360 pKa = 3.6YY361 pKa = 11.25LKK363 pKa = 10.03TIQHH367 pKa = 6.55ANHH370 pKa = 6.98MINLNSPIHH379 pKa = 6.59NDD381 pKa = 2.91WQLLIDD387 pKa = 5.67DD388 pKa = 4.4DD389 pKa = 4.46TALKK393 pKa = 11.02LMTPQLKK400 pKa = 9.91RR401 pKa = 11.84IYY403 pKa = 10.05HH404 pKa = 5.45KK405 pKa = 9.64QHH407 pKa = 6.32SNRR410 pKa = 11.84VYY412 pKa = 8.5TWDD415 pKa = 3.55YY416 pKa = 8.28LTPSFIQAYY425 pKa = 9.07PNYY428 pKa = 8.4KK429 pKa = 8.46TRR431 pKa = 11.84KK432 pKa = 9.31KK433 pKa = 9.92PFPLARR439 pKa = 11.84TIPQQRR445 pKa = 11.84QLLINHH451 pKa = 5.96NTRR454 pKa = 11.84EE455 pKa = 4.37LITLL459 pKa = 4.29
MM1 pKa = 6.87EE2 pKa = 4.95TVPGIITNHH11 pKa = 4.84NRR13 pKa = 11.84LLRR16 pKa = 11.84RR17 pKa = 11.84TLYY20 pKa = 10.36HH21 pKa = 6.13PPIYY25 pKa = 10.3APHH28 pKa = 7.12DD29 pKa = 3.95FYY31 pKa = 11.79YY32 pKa = 10.17YY33 pKa = 10.46AQSATTTIAAYY44 pKa = 10.07HH45 pKa = 5.67NRR47 pKa = 11.84HH48 pKa = 5.94RR49 pKa = 11.84PIILPDD55 pKa = 3.48YY56 pKa = 10.25NPSHH60 pKa = 6.91FNVDD64 pKa = 3.53NIFDD68 pKa = 3.69KK69 pKa = 10.97FKK71 pKa = 11.02RR72 pKa = 11.84RR73 pKa = 11.84LQPWTFEE80 pKa = 4.14QYY82 pKa = 10.08ISSMDD87 pKa = 3.45SPRR90 pKa = 11.84KK91 pKa = 9.33RR92 pKa = 11.84KK93 pKa = 9.4FYY95 pKa = 10.56QDD97 pKa = 3.59AYY99 pKa = 9.98TSILQGRR106 pKa = 11.84KK107 pKa = 6.77TSSGITPFTKK117 pKa = 10.25LEE119 pKa = 3.97KK120 pKa = 10.05MSTSKK125 pKa = 11.12YY126 pKa = 9.02KK127 pKa = 10.62APRR130 pKa = 11.84LIQGRR135 pKa = 11.84HH136 pKa = 3.97PTFNLCYY143 pKa = 10.14GRR145 pKa = 11.84YY146 pKa = 8.84IKK148 pKa = 10.4PLEE151 pKa = 4.32RR152 pKa = 11.84SLKK155 pKa = 9.73GDD157 pKa = 3.28RR158 pKa = 11.84HH159 pKa = 5.76FGKK162 pKa = 9.03GTYY165 pKa = 10.34DD166 pKa = 3.93DD167 pKa = 4.1IGRR170 pKa = 11.84KK171 pKa = 7.15IHH173 pKa = 6.35KK174 pKa = 10.12LSRR177 pKa = 11.84KK178 pKa = 7.88YY179 pKa = 10.51KK180 pKa = 8.49YY181 pKa = 9.07YY182 pKa = 10.7TEE184 pKa = 5.02GDD186 pKa = 3.3HH187 pKa = 5.74TTFDD191 pKa = 3.49SHH193 pKa = 5.5VTVEE197 pKa = 4.04MLRR200 pKa = 11.84LCHH203 pKa = 6.7RR204 pKa = 11.84FYY206 pKa = 11.23LRR208 pKa = 11.84CYY210 pKa = 8.62KK211 pKa = 10.29QSPEE215 pKa = 4.27LLKK218 pKa = 10.88LCKK221 pKa = 9.35KK222 pKa = 8.68TIRR225 pKa = 11.84NRR227 pKa = 11.84AVARR231 pKa = 11.84NGEE234 pKa = 4.39KK235 pKa = 9.42YY236 pKa = 7.83TVRR239 pKa = 11.84GTRR242 pKa = 11.84MSGDD246 pKa = 3.12VDD248 pKa = 3.51TGFGNSIINYY258 pKa = 8.11YY259 pKa = 9.81IIRR262 pKa = 11.84HH263 pKa = 4.72ALKK266 pKa = 10.19RR267 pKa = 11.84LHH269 pKa = 6.9IDD271 pKa = 2.92GDD273 pKa = 4.42AIVNGDD279 pKa = 3.89DD280 pKa = 5.52FIIFTNIKK288 pKa = 9.62ISTAEE293 pKa = 3.74FTNLLRR299 pKa = 11.84EE300 pKa = 4.34YY301 pKa = 11.41NMEE304 pKa = 4.07TTLGNSVDD312 pKa = 4.58SIHH315 pKa = 6.22QVEE318 pKa = 5.49FCRR321 pKa = 11.84CRR323 pKa = 11.84LVYY326 pKa = 10.5HH327 pKa = 7.16PDD329 pKa = 3.02GHH331 pKa = 6.34PTMAFDD337 pKa = 3.46PHH339 pKa = 8.13RR340 pKa = 11.84LEE342 pKa = 5.61KK343 pKa = 10.19IYY345 pKa = 10.86GCSNILYY352 pKa = 7.75PKK354 pKa = 8.93KK355 pKa = 10.35KK356 pKa = 10.21KK357 pKa = 10.82YY358 pKa = 10.31EE359 pKa = 4.49DD360 pKa = 3.6YY361 pKa = 11.25LKK363 pKa = 10.03TIQHH367 pKa = 6.55ANHH370 pKa = 6.98MINLNSPIHH379 pKa = 6.59NDD381 pKa = 2.91WQLLIDD387 pKa = 5.67DD388 pKa = 4.4DD389 pKa = 4.46TALKK393 pKa = 11.02LMTPQLKK400 pKa = 9.91RR401 pKa = 11.84IYY403 pKa = 10.05HH404 pKa = 5.45KK405 pKa = 9.64QHH407 pKa = 6.32SNRR410 pKa = 11.84VYY412 pKa = 8.5TWDD415 pKa = 3.55YY416 pKa = 8.28LTPSFIQAYY425 pKa = 9.07PNYY428 pKa = 8.4KK429 pKa = 8.46TRR431 pKa = 11.84KK432 pKa = 9.31KK433 pKa = 9.92PFPLARR439 pKa = 11.84TIPQQRR445 pKa = 11.84QLLINHH451 pKa = 5.96NTRR454 pKa = 11.84EE455 pKa = 4.37LITLL459 pKa = 4.29
Molecular weight: 54.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1505 |
459 |
553 |
501.7 |
57.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.648 ± 0.908 | 1.927 ± 0.444 |
5.249 ± 0.173 | 3.522 ± 0.662 |
4.186 ± 0.677 | 3.056 ± 0.629 |
3.189 ± 0.775 | 7.708 ± 0.676 |
4.983 ± 0.99 | 9.568 ± 0.61 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.86 ± 0.093 | 4.85 ± 0.269 |
5.847 ± 0.594 | 4.718 ± 0.618 |
5.847 ± 0.869 | 7.442 ± 1.025 |
9.369 ± 0.647 | 3.787 ± 0.537 |
0.997 ± 0.291 | 6.246 ± 0.43 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
