
Sewage-associated circular DNA virus-11
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.53
Get precalculated fractions of proteins
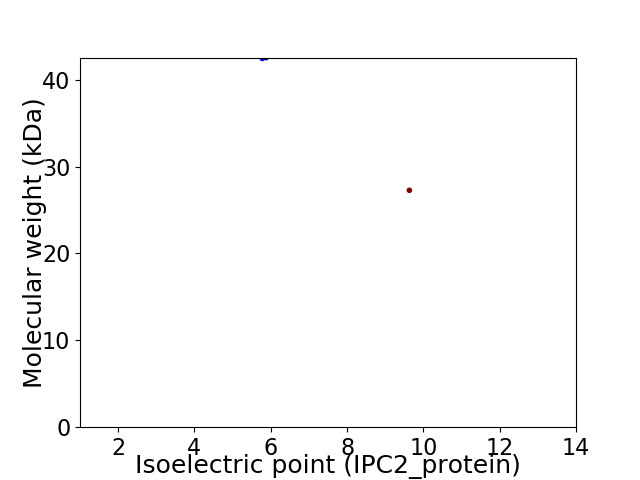
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075IZM2|A0A075IZM2_9VIRU ATP-dependent helicase Rep OS=Sewage-associated circular DNA virus-11 OX=1519387 PE=3 SV=1
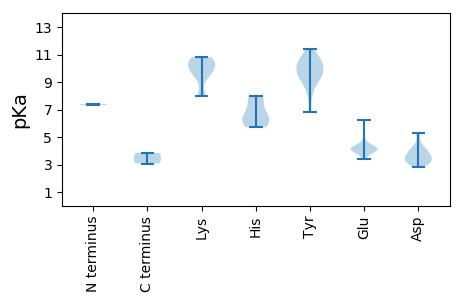
MM1 pKa = 7.39ATTTSSEE8 pKa = 3.91QSNKK12 pKa = 9.56SNRR15 pKa = 11.84TKK17 pKa = 10.5RR18 pKa = 11.84LSRR21 pKa = 11.84FVFTLNNYY29 pKa = 6.85TAEE32 pKa = 4.14EE33 pKa = 4.05EE34 pKa = 4.55QVIQLLGTTLKK45 pKa = 10.07MKK47 pKa = 9.33WLIYY51 pKa = 9.76GRR53 pKa = 11.84EE54 pKa = 4.13VGEE57 pKa = 4.45KK58 pKa = 8.96GTPHH62 pKa = 6.2LQGAAVIGRR71 pKa = 11.84QLAWKK76 pKa = 7.96TIKK79 pKa = 10.66NSFPPRR85 pKa = 11.84THH87 pKa = 7.62LEE89 pKa = 4.1EE90 pKa = 4.2MKK92 pKa = 9.84GTPRR96 pKa = 11.84EE97 pKa = 4.04SFIYY101 pKa = 8.64CTKK104 pKa = 10.33EE105 pKa = 3.56DD106 pKa = 3.3KK107 pKa = 10.46EE108 pKa = 4.4YY109 pKa = 10.5YY110 pKa = 10.05QFGEE114 pKa = 4.1EE115 pKa = 4.14PKK117 pKa = 10.03PGKK120 pKa = 10.63RR121 pKa = 11.84NDD123 pKa = 3.92LLDD126 pKa = 3.85AVASLRR132 pKa = 11.84KK133 pKa = 8.09TSSFRR138 pKa = 11.84DD139 pKa = 4.03FVKK142 pKa = 10.52DD143 pKa = 3.56DD144 pKa = 3.7QNASVYY150 pKa = 10.4VKK152 pKa = 7.98YY153 pKa = 9.42TRR155 pKa = 11.84GLTAFRR161 pKa = 11.84SLLRR165 pKa = 11.84EE166 pKa = 3.88PRR168 pKa = 11.84SPSRR172 pKa = 11.84PPTVYY177 pKa = 9.6WLHH180 pKa = 6.46GPTGAGKK187 pKa = 8.08TKK189 pKa = 10.3HH190 pKa = 5.74AWEE193 pKa = 4.07FLEE196 pKa = 6.21GYY198 pKa = 10.15FGTDD202 pKa = 2.83QEE204 pKa = 4.68TWISNGSLRR213 pKa = 11.84WFDD216 pKa = 4.27GYY218 pKa = 11.02DD219 pKa = 3.24GHH221 pKa = 7.99RR222 pKa = 11.84GVILDD227 pKa = 5.12DD228 pKa = 3.37IRR230 pKa = 11.84ADD232 pKa = 3.41SVPYY236 pKa = 10.76NEE238 pKa = 5.5LLRR241 pKa = 11.84LLDD244 pKa = 4.16RR245 pKa = 11.84YY246 pKa = 9.74PYY248 pKa = 9.57RR249 pKa = 11.84VEE251 pKa = 4.14YY252 pKa = 10.65KK253 pKa = 10.64GGTTEE258 pKa = 5.05WIPEE262 pKa = 4.2IIIITAPMGPYY273 pKa = 9.85QFFRR277 pKa = 11.84VGDD280 pKa = 4.52DD281 pKa = 3.01IEE283 pKa = 4.21QLLRR287 pKa = 11.84RR288 pKa = 11.84VNGGIFEE295 pKa = 3.99FTIDD299 pKa = 3.77GPNPRR304 pKa = 11.84LQTVAPTVPNTPAHH318 pKa = 6.35SEE320 pKa = 4.18VEE322 pKa = 4.38SQDD325 pKa = 4.01DD326 pKa = 3.82VQEE329 pKa = 4.19LQEE332 pKa = 4.25SSEE335 pKa = 4.08GLSLLEE341 pKa = 4.29SIGRR345 pKa = 11.84GSGIEE350 pKa = 3.8QDD352 pKa = 3.29QSNSQEE358 pKa = 4.19EE359 pKa = 4.34EE360 pKa = 3.78TQLRR364 pKa = 11.84KK365 pKa = 9.55KK366 pKa = 10.63RR367 pKa = 11.84KK368 pKa = 9.75FIDD371 pKa = 3.07
MM1 pKa = 7.39ATTTSSEE8 pKa = 3.91QSNKK12 pKa = 9.56SNRR15 pKa = 11.84TKK17 pKa = 10.5RR18 pKa = 11.84LSRR21 pKa = 11.84FVFTLNNYY29 pKa = 6.85TAEE32 pKa = 4.14EE33 pKa = 4.05EE34 pKa = 4.55QVIQLLGTTLKK45 pKa = 10.07MKK47 pKa = 9.33WLIYY51 pKa = 9.76GRR53 pKa = 11.84EE54 pKa = 4.13VGEE57 pKa = 4.45KK58 pKa = 8.96GTPHH62 pKa = 6.2LQGAAVIGRR71 pKa = 11.84QLAWKK76 pKa = 7.96TIKK79 pKa = 10.66NSFPPRR85 pKa = 11.84THH87 pKa = 7.62LEE89 pKa = 4.1EE90 pKa = 4.2MKK92 pKa = 9.84GTPRR96 pKa = 11.84EE97 pKa = 4.04SFIYY101 pKa = 8.64CTKK104 pKa = 10.33EE105 pKa = 3.56DD106 pKa = 3.3KK107 pKa = 10.46EE108 pKa = 4.4YY109 pKa = 10.5YY110 pKa = 10.05QFGEE114 pKa = 4.1EE115 pKa = 4.14PKK117 pKa = 10.03PGKK120 pKa = 10.63RR121 pKa = 11.84NDD123 pKa = 3.92LLDD126 pKa = 3.85AVASLRR132 pKa = 11.84KK133 pKa = 8.09TSSFRR138 pKa = 11.84DD139 pKa = 4.03FVKK142 pKa = 10.52DD143 pKa = 3.56DD144 pKa = 3.7QNASVYY150 pKa = 10.4VKK152 pKa = 7.98YY153 pKa = 9.42TRR155 pKa = 11.84GLTAFRR161 pKa = 11.84SLLRR165 pKa = 11.84EE166 pKa = 3.88PRR168 pKa = 11.84SPSRR172 pKa = 11.84PPTVYY177 pKa = 9.6WLHH180 pKa = 6.46GPTGAGKK187 pKa = 8.08TKK189 pKa = 10.3HH190 pKa = 5.74AWEE193 pKa = 4.07FLEE196 pKa = 6.21GYY198 pKa = 10.15FGTDD202 pKa = 2.83QEE204 pKa = 4.68TWISNGSLRR213 pKa = 11.84WFDD216 pKa = 4.27GYY218 pKa = 11.02DD219 pKa = 3.24GHH221 pKa = 7.99RR222 pKa = 11.84GVILDD227 pKa = 5.12DD228 pKa = 3.37IRR230 pKa = 11.84ADD232 pKa = 3.41SVPYY236 pKa = 10.76NEE238 pKa = 5.5LLRR241 pKa = 11.84LLDD244 pKa = 4.16RR245 pKa = 11.84YY246 pKa = 9.74PYY248 pKa = 9.57RR249 pKa = 11.84VEE251 pKa = 4.14YY252 pKa = 10.65KK253 pKa = 10.64GGTTEE258 pKa = 5.05WIPEE262 pKa = 4.2IIIITAPMGPYY273 pKa = 9.85QFFRR277 pKa = 11.84VGDD280 pKa = 4.52DD281 pKa = 3.01IEE283 pKa = 4.21QLLRR287 pKa = 11.84RR288 pKa = 11.84VNGGIFEE295 pKa = 3.99FTIDD299 pKa = 3.77GPNPRR304 pKa = 11.84LQTVAPTVPNTPAHH318 pKa = 6.35SEE320 pKa = 4.18VEE322 pKa = 4.38SQDD325 pKa = 4.01DD326 pKa = 3.82VQEE329 pKa = 4.19LQEE332 pKa = 4.25SSEE335 pKa = 4.08GLSLLEE341 pKa = 4.29SIGRR345 pKa = 11.84GSGIEE350 pKa = 3.8QDD352 pKa = 3.29QSNSQEE358 pKa = 4.19EE359 pKa = 4.34EE360 pKa = 3.78TQLRR364 pKa = 11.84KK365 pKa = 9.55KK366 pKa = 10.63RR367 pKa = 11.84KK368 pKa = 9.75FIDD371 pKa = 3.07
Molecular weight: 42.45 kDa
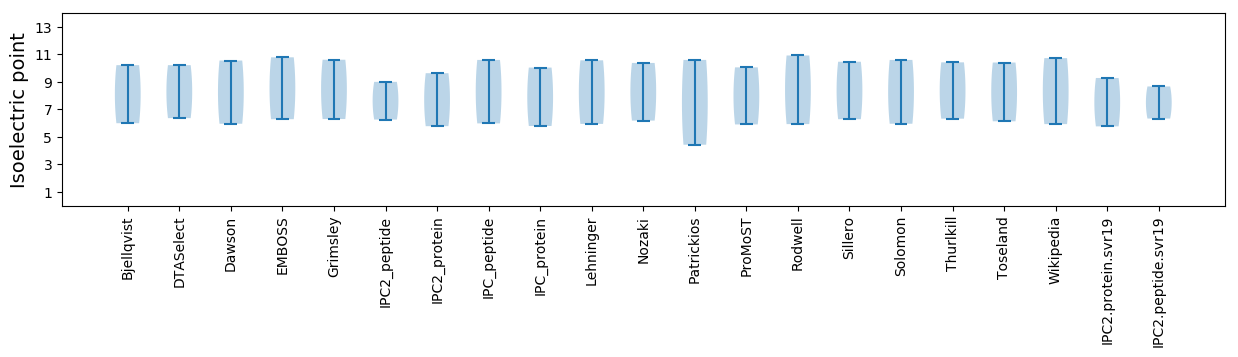
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075IZM2|A0A075IZM2_9VIRU ATP-dependent helicase Rep OS=Sewage-associated circular DNA virus-11 OX=1519387 PE=3 SV=1
MM1 pKa = 7.31KK2 pKa = 9.74RR3 pKa = 11.84QRR5 pKa = 11.84SAYY8 pKa = 9.16EE9 pKa = 3.41IEE11 pKa = 4.07NRR13 pKa = 11.84KK14 pKa = 8.02QAARR18 pKa = 11.84AKK20 pKa = 10.09RR21 pKa = 11.84RR22 pKa = 11.84ALVAARR28 pKa = 11.84AAPMRR33 pKa = 11.84AYY35 pKa = 10.48ANSQVPLRR43 pKa = 11.84SGGYY47 pKa = 9.75RR48 pKa = 11.84FGSTEE53 pKa = 3.83KK54 pKa = 10.74KK55 pKa = 10.8VFDD58 pKa = 5.26LDD60 pKa = 3.77TGTTNINTTGSVLALFVPTLGTDD83 pKa = 2.99MTNRR87 pKa = 11.84IGRR90 pKa = 11.84KK91 pKa = 9.36AIIKK95 pKa = 9.64SFYY98 pKa = 8.99LRR100 pKa = 11.84GYY102 pKa = 7.98VRR104 pKa = 11.84CEE106 pKa = 3.57NSLTPTSPQAGSSQQLRR123 pKa = 11.84IIVLIDD129 pKa = 3.36MQPNGALPAITDD141 pKa = 3.47ILKK144 pKa = 9.89EE145 pKa = 3.99QFPSSQLNINNRR157 pKa = 11.84DD158 pKa = 3.12RR159 pKa = 11.84FKK161 pKa = 10.69ILKK164 pKa = 9.84DD165 pKa = 2.97KK166 pKa = 10.77TYY168 pKa = 11.39ALDD171 pKa = 3.43TFIYY175 pKa = 8.99NTTATQAVGVAGRR188 pKa = 11.84TLQNFKK194 pKa = 10.5CYY196 pKa = 10.57KK197 pKa = 9.9KK198 pKa = 10.23MNLEE202 pKa = 4.29VIFNSGNAGTIADD215 pKa = 4.27TQTGALLIATIGSTASGTADD235 pKa = 3.16GNLVMSSRR243 pKa = 11.84IRR245 pKa = 11.84FLDD248 pKa = 3.39PP249 pKa = 3.84
MM1 pKa = 7.31KK2 pKa = 9.74RR3 pKa = 11.84QRR5 pKa = 11.84SAYY8 pKa = 9.16EE9 pKa = 3.41IEE11 pKa = 4.07NRR13 pKa = 11.84KK14 pKa = 8.02QAARR18 pKa = 11.84AKK20 pKa = 10.09RR21 pKa = 11.84RR22 pKa = 11.84ALVAARR28 pKa = 11.84AAPMRR33 pKa = 11.84AYY35 pKa = 10.48ANSQVPLRR43 pKa = 11.84SGGYY47 pKa = 9.75RR48 pKa = 11.84FGSTEE53 pKa = 3.83KK54 pKa = 10.74KK55 pKa = 10.8VFDD58 pKa = 5.26LDD60 pKa = 3.77TGTTNINTTGSVLALFVPTLGTDD83 pKa = 2.99MTNRR87 pKa = 11.84IGRR90 pKa = 11.84KK91 pKa = 9.36AIIKK95 pKa = 9.64SFYY98 pKa = 8.99LRR100 pKa = 11.84GYY102 pKa = 7.98VRR104 pKa = 11.84CEE106 pKa = 3.57NSLTPTSPQAGSSQQLRR123 pKa = 11.84IIVLIDD129 pKa = 3.36MQPNGALPAITDD141 pKa = 3.47ILKK144 pKa = 9.89EE145 pKa = 3.99QFPSSQLNINNRR157 pKa = 11.84DD158 pKa = 3.12RR159 pKa = 11.84FKK161 pKa = 10.69ILKK164 pKa = 9.84DD165 pKa = 2.97KK166 pKa = 10.77TYY168 pKa = 11.39ALDD171 pKa = 3.43TFIYY175 pKa = 8.99NTTATQAVGVAGRR188 pKa = 11.84TLQNFKK194 pKa = 10.5CYY196 pKa = 10.57KK197 pKa = 9.9KK198 pKa = 10.23MNLEE202 pKa = 4.29VIFNSGNAGTIADD215 pKa = 4.27TQTGALLIATIGSTASGTADD235 pKa = 3.16GNLVMSSRR243 pKa = 11.84IRR245 pKa = 11.84FLDD248 pKa = 3.39PP249 pKa = 3.84
Molecular weight: 27.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
620 |
249 |
371 |
310.0 |
34.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.613 ± 2.135 | 0.484 ± 0.178 |
5.0 ± 0.325 | 6.29 ± 2.164 |
4.194 ± 0.099 | 7.742 ± 0.286 |
0.968 ± 0.54 | 5.968 ± 0.703 |
5.645 ± 0.013 | 8.387 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.613 ± 0.444 | 4.677 ± 0.975 |
4.839 ± 0.683 | 4.839 ± 0.011 |
7.742 ± 0.162 | 6.935 ± 0.06 |
8.548 ± 0.608 | 4.677 ± 0.145 |
1.129 ± 0.63 | 3.71 ± 0.277 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
