
Flavobacterium beibuense F44-8
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavobacterium; Flavobacterium beibuense
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
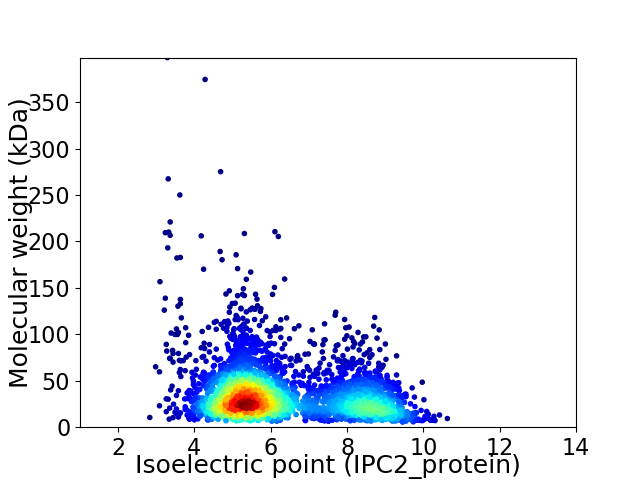
Virtual 2D-PAGE plot for 3263 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A2LWF6|A0A0A2LWF6_9FLAO Peptidase M22 OS=Flavobacterium beibuense F44-8 OX=1406840 GN=Q763_10825 PE=4 SV=1
MM1 pKa = 7.46KK2 pKa = 10.52KK3 pKa = 10.22KK4 pKa = 10.46LLYY7 pKa = 10.77LLLLFSGIASAQIVNIPDD25 pKa = 3.55ANFKK29 pKa = 11.05NKK31 pKa = 9.8LLSANTTNNIAKK43 pKa = 9.95DD44 pKa = 3.83EE45 pKa = 4.38NGVAIKK51 pKa = 9.98IDD53 pKa = 3.45QNNDD57 pKa = 3.02YY58 pKa = 10.38EE59 pKa = 4.44IQEE62 pKa = 4.28SEE64 pKa = 4.36ALLVWEE70 pKa = 5.33LSLFSAGMADD80 pKa = 3.42ATGLEE85 pKa = 4.68YY86 pKa = 9.32FTNLRR91 pKa = 11.84SLEE94 pKa = 4.12CANNSDD100 pKa = 5.25LVYY103 pKa = 10.86LDD105 pKa = 4.69LSSLINLEE113 pKa = 5.09DD114 pKa = 4.31FNCDD118 pKa = 2.73GGTGLGFLSFSGLFNLQTVYY138 pKa = 10.67ISDD141 pKa = 4.02TNLEE145 pKa = 4.46TIDD148 pKa = 3.63LTGVTGVQEE157 pKa = 4.48FEE159 pKa = 4.51LEE161 pKa = 4.33GVPVTSLDD169 pKa = 3.51FSDD172 pKa = 4.68MINLYY177 pKa = 10.63SLALSGTEE185 pKa = 5.06LVNLDD190 pKa = 4.54LGDD193 pKa = 4.54APLVTYY199 pKa = 10.73INLSYY204 pKa = 11.43NDD206 pKa = 3.62VLEE209 pKa = 4.75TISIKK214 pKa = 10.7NGAIIPSPSNIHH226 pKa = 6.52INNNNPQLNFICIDD240 pKa = 3.73EE241 pKa = 5.21DD242 pKa = 3.6EE243 pKa = 4.86LSQIEE248 pKa = 4.36LVDD251 pKa = 4.7GIEE254 pKa = 4.48NIVVSTYY261 pKa = 9.99CTFTPGGDD269 pKa = 3.52YY270 pKa = 10.19NTITGTAVFDD280 pKa = 4.67ADD282 pKa = 4.23SDD284 pKa = 4.3GCGPEE289 pKa = 4.27DD290 pKa = 3.84FPYY293 pKa = 10.71PFVKK297 pKa = 10.7LSINDD302 pKa = 3.93GVEE305 pKa = 3.57GGYY308 pKa = 8.37TFTGVNGEE316 pKa = 4.19YY317 pKa = 10.57SFYY320 pKa = 10.27TQDD323 pKa = 3.02GNFTVVPEE331 pKa = 5.02FEE333 pKa = 5.75DD334 pKa = 2.96NWFTVTPAYY343 pKa = 11.24VNFPVVDD350 pKa = 3.72NSVSVQNFCIEE361 pKa = 4.26PNGVHH366 pKa = 6.57NDD368 pKa = 3.43VEE370 pKa = 4.83VVMVPITPARR380 pKa = 11.84PGFDD384 pKa = 3.87AIYY387 pKa = 10.33KK388 pKa = 9.78IVYY391 pKa = 8.82KK392 pKa = 10.95NKK394 pKa = 9.18GNQVVSGTVGCEE406 pKa = 3.38WDD408 pKa = 4.13YY409 pKa = 11.12EE410 pKa = 4.53TLTPLVLNPMADD422 pKa = 4.07DD423 pKa = 4.08IAPGVYY429 pKa = 8.98GWNYY433 pKa = 10.15TNLQPFEE440 pKa = 4.06NRR442 pKa = 11.84EE443 pKa = 3.76ILMTLHH449 pKa = 6.4VNSPTDD455 pKa = 3.57TPAVNIDD462 pKa = 3.56DD463 pKa = 4.3VLPFTANIDD472 pKa = 3.43GGTDD476 pKa = 3.26EE477 pKa = 5.41NPNNNQYY484 pKa = 10.4IFNQTVVGSFDD495 pKa = 4.2PNNIICIEE503 pKa = 4.12GEE505 pKa = 4.38TLPSNAIGDD514 pKa = 3.91YY515 pKa = 10.52LHH517 pKa = 6.64YY518 pKa = 10.77VVNFEE523 pKa = 4.34NTGTAPASFVVVKK536 pKa = 9.67HH537 pKa = 6.13VINEE541 pKa = 3.77ADD543 pKa = 3.32FDD545 pKa = 4.64INSIQILNSSHH556 pKa = 5.18QVEE559 pKa = 5.4ARR561 pKa = 11.84IQDD564 pKa = 3.6NVIEE568 pKa = 5.06FIFDD572 pKa = 4.07NINLNAADD580 pKa = 4.26HH581 pKa = 6.3GNILFKK587 pKa = 10.98LKK589 pKa = 10.5SKK591 pKa = 10.93QDD593 pKa = 3.59LQEE596 pKa = 4.24GDD598 pKa = 3.35NVMNEE603 pKa = 3.33AGIYY607 pKa = 9.84FDD609 pKa = 4.53YY610 pKa = 10.86NLPVATNQAVTEE622 pKa = 4.42FEE624 pKa = 5.69DD625 pKa = 5.13IMGLDD630 pKa = 5.11DD631 pKa = 5.4FIKK634 pKa = 10.86DD635 pKa = 3.57NSIKK639 pKa = 10.59VYY641 pKa = 10.17PNPAKK646 pKa = 10.54DD647 pKa = 3.63VLSVKK652 pKa = 10.59GDD654 pKa = 3.35VNLRR658 pKa = 11.84SVSLYY663 pKa = 10.46DD664 pKa = 3.16IQGRR668 pKa = 11.84LLQTVMPNDD677 pKa = 3.51VQVTIDD683 pKa = 2.72IAMRR687 pKa = 11.84AKK689 pKa = 10.53GIYY692 pKa = 8.75FLKK695 pKa = 9.27VTSDD699 pKa = 3.15KK700 pKa = 10.9GVKK703 pKa = 8.68VEE705 pKa = 4.6KK706 pKa = 10.46VIKK709 pKa = 9.81EE710 pKa = 3.93
MM1 pKa = 7.46KK2 pKa = 10.52KK3 pKa = 10.22KK4 pKa = 10.46LLYY7 pKa = 10.77LLLLFSGIASAQIVNIPDD25 pKa = 3.55ANFKK29 pKa = 11.05NKK31 pKa = 9.8LLSANTTNNIAKK43 pKa = 9.95DD44 pKa = 3.83EE45 pKa = 4.38NGVAIKK51 pKa = 9.98IDD53 pKa = 3.45QNNDD57 pKa = 3.02YY58 pKa = 10.38EE59 pKa = 4.44IQEE62 pKa = 4.28SEE64 pKa = 4.36ALLVWEE70 pKa = 5.33LSLFSAGMADD80 pKa = 3.42ATGLEE85 pKa = 4.68YY86 pKa = 9.32FTNLRR91 pKa = 11.84SLEE94 pKa = 4.12CANNSDD100 pKa = 5.25LVYY103 pKa = 10.86LDD105 pKa = 4.69LSSLINLEE113 pKa = 5.09DD114 pKa = 4.31FNCDD118 pKa = 2.73GGTGLGFLSFSGLFNLQTVYY138 pKa = 10.67ISDD141 pKa = 4.02TNLEE145 pKa = 4.46TIDD148 pKa = 3.63LTGVTGVQEE157 pKa = 4.48FEE159 pKa = 4.51LEE161 pKa = 4.33GVPVTSLDD169 pKa = 3.51FSDD172 pKa = 4.68MINLYY177 pKa = 10.63SLALSGTEE185 pKa = 5.06LVNLDD190 pKa = 4.54LGDD193 pKa = 4.54APLVTYY199 pKa = 10.73INLSYY204 pKa = 11.43NDD206 pKa = 3.62VLEE209 pKa = 4.75TISIKK214 pKa = 10.7NGAIIPSPSNIHH226 pKa = 6.52INNNNPQLNFICIDD240 pKa = 3.73EE241 pKa = 5.21DD242 pKa = 3.6EE243 pKa = 4.86LSQIEE248 pKa = 4.36LVDD251 pKa = 4.7GIEE254 pKa = 4.48NIVVSTYY261 pKa = 9.99CTFTPGGDD269 pKa = 3.52YY270 pKa = 10.19NTITGTAVFDD280 pKa = 4.67ADD282 pKa = 4.23SDD284 pKa = 4.3GCGPEE289 pKa = 4.27DD290 pKa = 3.84FPYY293 pKa = 10.71PFVKK297 pKa = 10.7LSINDD302 pKa = 3.93GVEE305 pKa = 3.57GGYY308 pKa = 8.37TFTGVNGEE316 pKa = 4.19YY317 pKa = 10.57SFYY320 pKa = 10.27TQDD323 pKa = 3.02GNFTVVPEE331 pKa = 5.02FEE333 pKa = 5.75DD334 pKa = 2.96NWFTVTPAYY343 pKa = 11.24VNFPVVDD350 pKa = 3.72NSVSVQNFCIEE361 pKa = 4.26PNGVHH366 pKa = 6.57NDD368 pKa = 3.43VEE370 pKa = 4.83VVMVPITPARR380 pKa = 11.84PGFDD384 pKa = 3.87AIYY387 pKa = 10.33KK388 pKa = 9.78IVYY391 pKa = 8.82KK392 pKa = 10.95NKK394 pKa = 9.18GNQVVSGTVGCEE406 pKa = 3.38WDD408 pKa = 4.13YY409 pKa = 11.12EE410 pKa = 4.53TLTPLVLNPMADD422 pKa = 4.07DD423 pKa = 4.08IAPGVYY429 pKa = 8.98GWNYY433 pKa = 10.15TNLQPFEE440 pKa = 4.06NRR442 pKa = 11.84EE443 pKa = 3.76ILMTLHH449 pKa = 6.4VNSPTDD455 pKa = 3.57TPAVNIDD462 pKa = 3.56DD463 pKa = 4.3VLPFTANIDD472 pKa = 3.43GGTDD476 pKa = 3.26EE477 pKa = 5.41NPNNNQYY484 pKa = 10.4IFNQTVVGSFDD495 pKa = 4.2PNNIICIEE503 pKa = 4.12GEE505 pKa = 4.38TLPSNAIGDD514 pKa = 3.91YY515 pKa = 10.52LHH517 pKa = 6.64YY518 pKa = 10.77VVNFEE523 pKa = 4.34NTGTAPASFVVVKK536 pKa = 9.67HH537 pKa = 6.13VINEE541 pKa = 3.77ADD543 pKa = 3.32FDD545 pKa = 4.64INSIQILNSSHH556 pKa = 5.18QVEE559 pKa = 5.4ARR561 pKa = 11.84IQDD564 pKa = 3.6NVIEE568 pKa = 5.06FIFDD572 pKa = 4.07NINLNAADD580 pKa = 4.26HH581 pKa = 6.3GNILFKK587 pKa = 10.98LKK589 pKa = 10.5SKK591 pKa = 10.93QDD593 pKa = 3.59LQEE596 pKa = 4.24GDD598 pKa = 3.35NVMNEE603 pKa = 3.33AGIYY607 pKa = 9.84FDD609 pKa = 4.53YY610 pKa = 10.86NLPVATNQAVTEE622 pKa = 4.42FEE624 pKa = 5.69DD625 pKa = 5.13IMGLDD630 pKa = 5.11DD631 pKa = 5.4FIKK634 pKa = 10.86DD635 pKa = 3.57NSIKK639 pKa = 10.59VYY641 pKa = 10.17PNPAKK646 pKa = 10.54DD647 pKa = 3.63VLSVKK652 pKa = 10.59GDD654 pKa = 3.35VNLRR658 pKa = 11.84SVSLYY663 pKa = 10.46DD664 pKa = 3.16IQGRR668 pKa = 11.84LLQTVMPNDD677 pKa = 3.51VQVTIDD683 pKa = 2.72IAMRR687 pKa = 11.84AKK689 pKa = 10.53GIYY692 pKa = 8.75FLKK695 pKa = 9.27VTSDD699 pKa = 3.15KK700 pKa = 10.9GVKK703 pKa = 8.68VEE705 pKa = 4.6KK706 pKa = 10.46VIKK709 pKa = 9.81EE710 pKa = 3.93
Molecular weight: 78.17 kDa
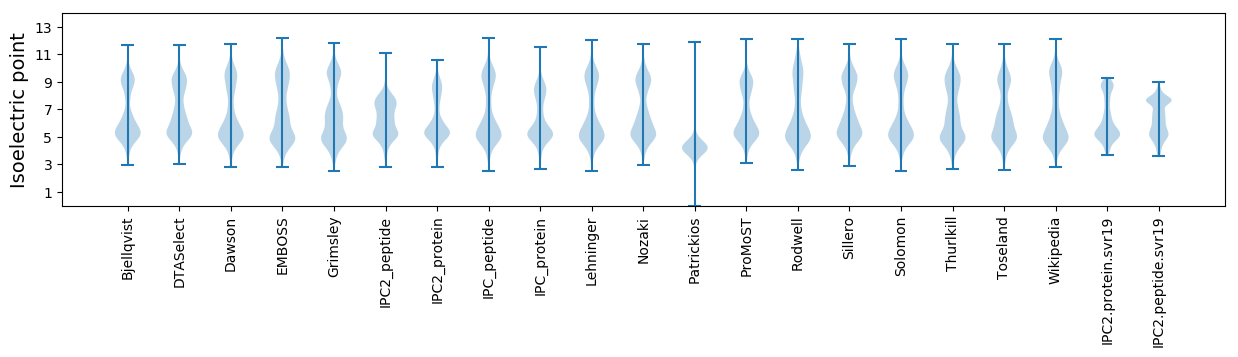
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A2LJ55|A0A0A2LJ55_9FLAO Transporter OS=Flavobacterium beibuense F44-8 OX=1406840 GN=Q763_11335 PE=3 SV=1
MM1 pKa = 7.5SFVTNIRR8 pKa = 11.84HH9 pKa = 4.78FFEE12 pKa = 5.25RR13 pKa = 11.84NGFHH17 pKa = 6.58VSARR21 pKa = 11.84LSDD24 pKa = 3.7RR25 pKa = 11.84LGMRR29 pKa = 11.84VTNVRR34 pKa = 11.84LFFIYY39 pKa = 10.23ISFFTVGLWFGVYY52 pKa = 8.36LTLAFWLRR60 pKa = 11.84LKK62 pKa = 10.75DD63 pKa = 3.42AVYY66 pKa = 9.69TKK68 pKa = 10.33RR69 pKa = 11.84SSVFDD74 pKa = 3.68LL75 pKa = 4.35
MM1 pKa = 7.5SFVTNIRR8 pKa = 11.84HH9 pKa = 4.78FFEE12 pKa = 5.25RR13 pKa = 11.84NGFHH17 pKa = 6.58VSARR21 pKa = 11.84LSDD24 pKa = 3.7RR25 pKa = 11.84LGMRR29 pKa = 11.84VTNVRR34 pKa = 11.84LFFIYY39 pKa = 10.23ISFFTVGLWFGVYY52 pKa = 8.36LTLAFWLRR60 pKa = 11.84LKK62 pKa = 10.75DD63 pKa = 3.42AVYY66 pKa = 9.69TKK68 pKa = 10.33RR69 pKa = 11.84SSVFDD74 pKa = 3.68LL75 pKa = 4.35
Molecular weight: 8.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1084350 |
46 |
3813 |
332.3 |
37.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.666 ± 0.049 | 0.811 ± 0.018 |
5.724 ± 0.034 | 6.748 ± 0.043 |
5.03 ± 0.039 | 6.491 ± 0.046 |
1.706 ± 0.021 | 7.408 ± 0.041 |
7.374 ± 0.079 | 8.915 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.337 ± 0.021 | 6.161 ± 0.045 |
3.504 ± 0.03 | 3.418 ± 0.024 |
3.361 ± 0.036 | 6.138 ± 0.037 |
6.198 ± 0.07 | 6.544 ± 0.042 |
1.036 ± 0.016 | 4.432 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
