
Pseudoruegeria sp. SK021
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Pseudoruegeria; unclassified Pseudoruegeria
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
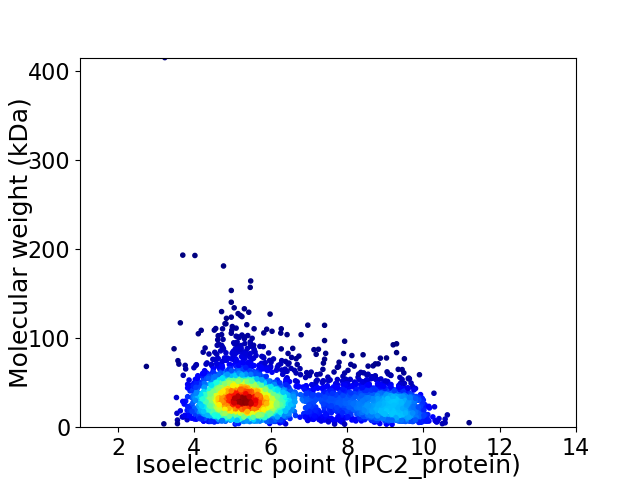
Virtual 2D-PAGE plot for 3609 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X4IUN9|A0A1X4IUN9_9RHOB Peptide ABC transporter permease OS=Pseudoruegeria sp. SK021 OX=1933035 GN=BV911_14265 PE=3 SV=1
MM1 pKa = 7.72SDD3 pKa = 3.26ILVKK7 pKa = 10.71SSSEE11 pKa = 3.74LVQALRR17 pKa = 11.84QATGGDD23 pKa = 4.03TIFLAPGDD31 pKa = 4.15YY32 pKa = 7.67EE33 pKa = 4.35TLFIKK38 pKa = 10.65DD39 pKa = 3.36LAYY42 pKa = 10.53ASDD45 pKa = 3.77VTLTSLDD52 pKa = 3.68AEE54 pKa = 4.37NPAVFTAPVTFVRR67 pKa = 11.84ANGLVLDD74 pKa = 4.72NIIFQSDD81 pKa = 4.16SIEE84 pKa = 4.27GTSGSAPLLFLHH96 pKa = 6.66SSEE99 pKa = 4.26NVTMTDD105 pKa = 3.69LVFTGYY111 pKa = 10.52LPSEE115 pKa = 4.4EE116 pKa = 4.68EE117 pKa = 4.27GVDD120 pKa = 3.31AYY122 pKa = 10.88AATTTRR128 pKa = 11.84GDD130 pKa = 3.85AITGYY135 pKa = 10.75GYY137 pKa = 8.36QTGLRR142 pKa = 11.84IHH144 pKa = 6.63NSDD147 pKa = 3.45NVSLEE152 pKa = 3.88DD153 pKa = 3.25ATFRR157 pKa = 11.84DD158 pKa = 3.86LRR160 pKa = 11.84YY161 pKa = 9.97AIYY164 pKa = 9.48TGEE167 pKa = 4.09STEE170 pKa = 3.92VDD172 pKa = 3.56LTGMSIEE179 pKa = 4.03QVRR182 pKa = 11.84EE183 pKa = 4.01GIMFYY188 pKa = 10.18EE189 pKa = 4.28VQGMVISDD197 pKa = 3.62SVFQNFKK204 pKa = 9.82PWLGMDD210 pKa = 3.79DD211 pKa = 3.85PAYY214 pKa = 8.93DD215 pKa = 4.22TNTHH219 pKa = 6.28DD220 pKa = 4.49HH221 pKa = 7.45PDD223 pKa = 3.75MIQYY227 pKa = 9.42WGDD230 pKa = 3.16NSTLGVHH237 pKa = 7.73DD238 pKa = 3.5ITITGNTFIAEE249 pKa = 4.43PGGSTQTIFGHH260 pKa = 7.11LGTGPAGVTASNFTVTDD277 pKa = 3.27NTIINGHH284 pKa = 5.69PHH286 pKa = 7.42AISLGDD292 pKa = 3.21VDD294 pKa = 4.47GAVISGNVLLPNSDD308 pKa = 3.77GLLKK312 pKa = 10.58YY313 pKa = 7.73WAPSIGAPGGQNLEE327 pKa = 4.06IYY329 pKa = 10.68DD330 pKa = 3.83NVVVYY335 pKa = 9.74WNGYY339 pKa = 7.2GVWALDD345 pKa = 3.62EE346 pKa = 4.22QEE348 pKa = 4.02RR349 pKa = 11.84AAANVSIGDD358 pKa = 3.76NTLLSDD364 pKa = 5.38DD365 pKa = 5.14PDD367 pKa = 5.05DD368 pKa = 3.9EE369 pKa = 5.0NYY371 pKa = 9.89WGFWSPASDD380 pKa = 3.77YY381 pKa = 11.39PLVQWAEE388 pKa = 3.92EE389 pKa = 4.07AGLGTVLGTEE399 pKa = 4.04TGEE402 pKa = 4.23VLWASSHH409 pKa = 5.64GSFIEE414 pKa = 4.11GLGGDD419 pKa = 3.87DD420 pKa = 3.92TLSGRR425 pKa = 11.84AGTDD429 pKa = 2.57ILTGGGGADD438 pKa = 3.01RR439 pKa = 11.84FLYY442 pKa = 10.5DD443 pKa = 3.12MRR445 pKa = 11.84GATVQGDD452 pKa = 4.27DD453 pKa = 5.66LILDD457 pKa = 4.33LDD459 pKa = 4.15FSEE462 pKa = 5.57GDD464 pKa = 3.24SLYY467 pKa = 10.56ILTGTMGLFSDD478 pKa = 4.68SAISGNDD485 pKa = 3.13LRR487 pKa = 11.84VLGDD491 pKa = 3.6GSSAIVDD498 pKa = 3.62SFADD502 pKa = 3.11IGEE505 pKa = 4.37LVASGAMIYY514 pKa = 10.73DD515 pKa = 3.43SDD517 pKa = 4.14GLGGGVLSLTSGDD530 pKa = 3.21GHH532 pKa = 7.46ALYY535 pKa = 10.23IAGLDD540 pKa = 3.6AASLPEE546 pKa = 4.17DD547 pKa = 4.43TIDD550 pKa = 3.93PTPADD555 pKa = 4.51PEE557 pKa = 4.15PDD559 pKa = 3.26ILGTAAGEE567 pKa = 4.28TLWASSRR574 pKa = 11.84GSFIDD579 pKa = 3.75ARR581 pKa = 11.84EE582 pKa = 4.06GDD584 pKa = 3.66DD585 pKa = 3.45TLFGRR590 pKa = 11.84TGADD594 pKa = 2.46ILMGGDD600 pKa = 3.21GADD603 pKa = 2.68RR604 pKa = 11.84FVFDD608 pKa = 5.09LRR610 pKa = 11.84SSSAAEE616 pKa = 4.29DD617 pKa = 4.53DD618 pKa = 5.64LILDD622 pKa = 4.56LDD624 pKa = 4.11FSEE627 pKa = 5.21GDD629 pKa = 3.49EE630 pKa = 4.9IYY632 pKa = 10.54ILTNTAGQFTDD643 pKa = 3.18SHH645 pKa = 7.23DD646 pKa = 4.03PGNDD650 pKa = 3.13LRR652 pKa = 11.84ILSDD656 pKa = 3.49GASVMITSLADD667 pKa = 3.56LQEE670 pKa = 4.46VVASGAMTYY679 pKa = 10.94DD680 pKa = 3.14SDD682 pKa = 4.07GDD684 pKa = 3.67SGGFLRR690 pKa = 11.84LADD693 pKa = 3.55GTGYY697 pKa = 11.17GLQIEE702 pKa = 4.74DD703 pKa = 5.41LYY705 pKa = 10.97PLLL708 pKa = 5.35
MM1 pKa = 7.72SDD3 pKa = 3.26ILVKK7 pKa = 10.71SSSEE11 pKa = 3.74LVQALRR17 pKa = 11.84QATGGDD23 pKa = 4.03TIFLAPGDD31 pKa = 4.15YY32 pKa = 7.67EE33 pKa = 4.35TLFIKK38 pKa = 10.65DD39 pKa = 3.36LAYY42 pKa = 10.53ASDD45 pKa = 3.77VTLTSLDD52 pKa = 3.68AEE54 pKa = 4.37NPAVFTAPVTFVRR67 pKa = 11.84ANGLVLDD74 pKa = 4.72NIIFQSDD81 pKa = 4.16SIEE84 pKa = 4.27GTSGSAPLLFLHH96 pKa = 6.66SSEE99 pKa = 4.26NVTMTDD105 pKa = 3.69LVFTGYY111 pKa = 10.52LPSEE115 pKa = 4.4EE116 pKa = 4.68EE117 pKa = 4.27GVDD120 pKa = 3.31AYY122 pKa = 10.88AATTTRR128 pKa = 11.84GDD130 pKa = 3.85AITGYY135 pKa = 10.75GYY137 pKa = 8.36QTGLRR142 pKa = 11.84IHH144 pKa = 6.63NSDD147 pKa = 3.45NVSLEE152 pKa = 3.88DD153 pKa = 3.25ATFRR157 pKa = 11.84DD158 pKa = 3.86LRR160 pKa = 11.84YY161 pKa = 9.97AIYY164 pKa = 9.48TGEE167 pKa = 4.09STEE170 pKa = 3.92VDD172 pKa = 3.56LTGMSIEE179 pKa = 4.03QVRR182 pKa = 11.84EE183 pKa = 4.01GIMFYY188 pKa = 10.18EE189 pKa = 4.28VQGMVISDD197 pKa = 3.62SVFQNFKK204 pKa = 9.82PWLGMDD210 pKa = 3.79DD211 pKa = 3.85PAYY214 pKa = 8.93DD215 pKa = 4.22TNTHH219 pKa = 6.28DD220 pKa = 4.49HH221 pKa = 7.45PDD223 pKa = 3.75MIQYY227 pKa = 9.42WGDD230 pKa = 3.16NSTLGVHH237 pKa = 7.73DD238 pKa = 3.5ITITGNTFIAEE249 pKa = 4.43PGGSTQTIFGHH260 pKa = 7.11LGTGPAGVTASNFTVTDD277 pKa = 3.27NTIINGHH284 pKa = 5.69PHH286 pKa = 7.42AISLGDD292 pKa = 3.21VDD294 pKa = 4.47GAVISGNVLLPNSDD308 pKa = 3.77GLLKK312 pKa = 10.58YY313 pKa = 7.73WAPSIGAPGGQNLEE327 pKa = 4.06IYY329 pKa = 10.68DD330 pKa = 3.83NVVVYY335 pKa = 9.74WNGYY339 pKa = 7.2GVWALDD345 pKa = 3.62EE346 pKa = 4.22QEE348 pKa = 4.02RR349 pKa = 11.84AAANVSIGDD358 pKa = 3.76NTLLSDD364 pKa = 5.38DD365 pKa = 5.14PDD367 pKa = 5.05DD368 pKa = 3.9EE369 pKa = 5.0NYY371 pKa = 9.89WGFWSPASDD380 pKa = 3.77YY381 pKa = 11.39PLVQWAEE388 pKa = 3.92EE389 pKa = 4.07AGLGTVLGTEE399 pKa = 4.04TGEE402 pKa = 4.23VLWASSHH409 pKa = 5.64GSFIEE414 pKa = 4.11GLGGDD419 pKa = 3.87DD420 pKa = 3.92TLSGRR425 pKa = 11.84AGTDD429 pKa = 2.57ILTGGGGADD438 pKa = 3.01RR439 pKa = 11.84FLYY442 pKa = 10.5DD443 pKa = 3.12MRR445 pKa = 11.84GATVQGDD452 pKa = 4.27DD453 pKa = 5.66LILDD457 pKa = 4.33LDD459 pKa = 4.15FSEE462 pKa = 5.57GDD464 pKa = 3.24SLYY467 pKa = 10.56ILTGTMGLFSDD478 pKa = 4.68SAISGNDD485 pKa = 3.13LRR487 pKa = 11.84VLGDD491 pKa = 3.6GSSAIVDD498 pKa = 3.62SFADD502 pKa = 3.11IGEE505 pKa = 4.37LVASGAMIYY514 pKa = 10.73DD515 pKa = 3.43SDD517 pKa = 4.14GLGGGVLSLTSGDD530 pKa = 3.21GHH532 pKa = 7.46ALYY535 pKa = 10.23IAGLDD540 pKa = 3.6AASLPEE546 pKa = 4.17DD547 pKa = 4.43TIDD550 pKa = 3.93PTPADD555 pKa = 4.51PEE557 pKa = 4.15PDD559 pKa = 3.26ILGTAAGEE567 pKa = 4.28TLWASSRR574 pKa = 11.84GSFIDD579 pKa = 3.75ARR581 pKa = 11.84EE582 pKa = 4.06GDD584 pKa = 3.66DD585 pKa = 3.45TLFGRR590 pKa = 11.84TGADD594 pKa = 2.46ILMGGDD600 pKa = 3.21GADD603 pKa = 2.68RR604 pKa = 11.84FVFDD608 pKa = 5.09LRR610 pKa = 11.84SSSAAEE616 pKa = 4.29DD617 pKa = 4.53DD618 pKa = 5.64LILDD622 pKa = 4.56LDD624 pKa = 4.11FSEE627 pKa = 5.21GDD629 pKa = 3.49EE630 pKa = 4.9IYY632 pKa = 10.54ILTNTAGQFTDD643 pKa = 3.18SHH645 pKa = 7.23DD646 pKa = 4.03PGNDD650 pKa = 3.13LRR652 pKa = 11.84ILSDD656 pKa = 3.49GASVMITSLADD667 pKa = 3.56LQEE670 pKa = 4.46VVASGAMTYY679 pKa = 10.94DD680 pKa = 3.14SDD682 pKa = 4.07GDD684 pKa = 3.67SGGFLRR690 pKa = 11.84LADD693 pKa = 3.55GTGYY697 pKa = 11.17GLQIEE702 pKa = 4.74DD703 pKa = 5.41LYY705 pKa = 10.97PLLL708 pKa = 5.35
Molecular weight: 74.71 kDa
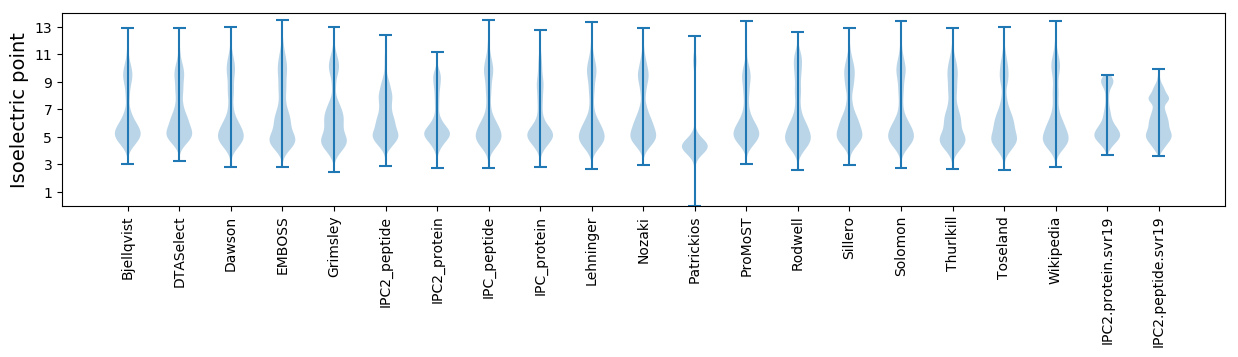
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X4J1T7|A0A1X4J1T7_9RHOB DUF4329 domain-containing protein OS=Pseudoruegeria sp. SK021 OX=1933035 GN=BV911_01190 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.82IINSRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.39GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.82IINSRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.39GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 5.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1135785 |
28 |
4002 |
314.7 |
34.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.259 ± 0.055 | 0.902 ± 0.013 |
6.215 ± 0.044 | 5.044 ± 0.032 |
3.756 ± 0.023 | 8.591 ± 0.038 |
2.006 ± 0.02 | 5.293 ± 0.033 |
2.849 ± 0.032 | 10.358 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.706 ± 0.02 | 2.544 ± 0.023 |
5.266 ± 0.027 | 3.298 ± 0.025 |
6.57 ± 0.046 | 5.483 ± 0.028 |
5.843 ± 0.039 | 7.432 ± 0.035 |
1.396 ± 0.017 | 2.189 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
