
Carassius auratus (Goldfish)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Actinopterygii; Actinopteri; Neopterygii; Teleostei; Osteoglossocephalai; Clupeocephala; Otomorpha; Ostariophysi<
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
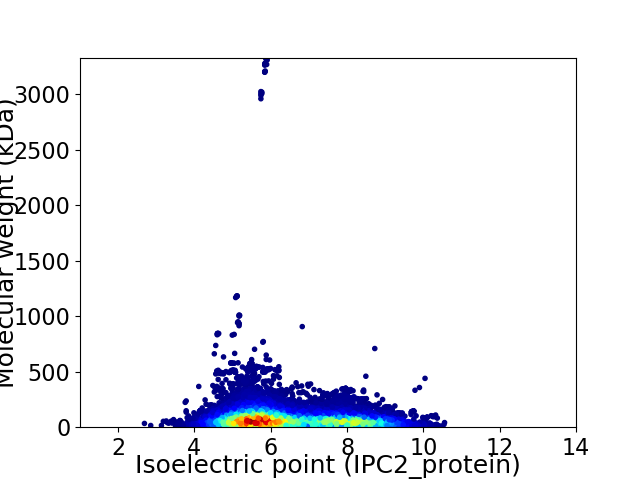
Virtual 2D-PAGE plot for 82965 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6P6LSX5|A0A6P6LSX5_CARAU LOW QUALITY PROTEIN: uncharacterized protein LOC113062173 OS=Carassius auratus OX=7957 GN=LOC113062173 PE=4 SV=1
MM1 pKa = 7.4LSSHH5 pKa = 5.8TFDD8 pKa = 4.7RR9 pKa = 11.84PAAGQTSEE17 pKa = 4.14GVGVVSSEE25 pKa = 4.27VQSQPTGSEE34 pKa = 3.79GFAPHH39 pKa = 6.91TDD41 pKa = 2.87GSQYY45 pKa = 11.31EE46 pKa = 4.37NGEE49 pKa = 4.02TAYY52 pKa = 10.47DD53 pKa = 4.24GSQIEE58 pKa = 4.45SPEE61 pKa = 3.79IPEE64 pKa = 4.5IEE66 pKa = 4.55SNGNGHH72 pKa = 6.38KK73 pKa = 10.23QLLNGGEE80 pKa = 4.19TGFTGVDD87 pKa = 2.9HH88 pKa = 6.9FMAGTSQIQEE98 pKa = 3.92GGGFDD103 pKa = 3.5SFGTSSHH110 pKa = 6.8LEE112 pKa = 3.91TTGVIDD118 pKa = 4.26QSSHH122 pKa = 6.76DD123 pKa = 3.95FLVDD127 pKa = 3.31LMGGIGEE134 pKa = 4.68SYY136 pKa = 11.15GPDD139 pKa = 3.3TQTDD143 pKa = 3.94GLGTLLDD150 pKa = 4.71LLDD153 pKa = 4.68TPPDD157 pKa = 3.67VPPTCLVTDD166 pKa = 4.48AVSTDD171 pKa = 2.89HH172 pKa = 6.74MGLAFQVDD180 pKa = 3.94SAAAGLSPGHH190 pKa = 6.91PSSGGPVLDD199 pKa = 5.32APPDD203 pKa = 4.04SPGLDD208 pKa = 3.3YY209 pKa = 11.6LFVDD213 pKa = 4.78NGNGDD218 pKa = 3.74YY219 pKa = 9.44THH221 pKa = 6.75SVSISDD227 pKa = 4.25NGAQSKK233 pKa = 10.69SPADD237 pKa = 3.69TTDD240 pKa = 2.91SSVVFDD246 pKa = 4.08TMSHH250 pKa = 6.95PDD252 pKa = 3.49HH253 pKa = 7.0FFTDD257 pKa = 4.41YY258 pKa = 11.3SDD260 pKa = 3.4GWVDD264 pKa = 4.17NNGADD269 pKa = 4.47LPEE272 pKa = 4.48TNGNGNGRR280 pKa = 11.84HH281 pKa = 5.55KK282 pKa = 10.62PVVDD286 pKa = 3.74MQKK289 pKa = 10.79GDD291 pKa = 3.69PQGVHH296 pKa = 6.67LDD298 pKa = 3.4ISGTGHH304 pKa = 6.28QDD306 pKa = 2.52AATAMHH312 pKa = 6.39HH313 pKa = 5.46TAVGALDD320 pKa = 5.28LNDD323 pKa = 4.14HH324 pKa = 5.91TLSPYY329 pKa = 10.54TDD331 pKa = 3.15TTGFDD336 pKa = 3.83GVIGAGAQTDD346 pKa = 3.72MAGAGGDD353 pKa = 3.94PATDD357 pKa = 3.68GQTLTDD363 pKa = 3.55MTGQGHH369 pKa = 7.02LAVTDD374 pKa = 3.57GMPNYY379 pKa = 9.25TADD382 pKa = 3.53SVRR385 pKa = 11.84ATGTGFTDD393 pKa = 4.13ASDD396 pKa = 3.61THH398 pKa = 8.17SSMVQTDD405 pKa = 4.1LPVTGDD411 pKa = 3.46PFTGVSSQTDD421 pKa = 3.08AMGTGQPGATEE432 pKa = 4.0QTQTAVSAGEE442 pKa = 4.04QYY444 pKa = 8.74HH445 pKa = 5.93TSGQGFEE452 pKa = 4.14GAEE455 pKa = 3.98NVEE458 pKa = 5.53LEE460 pKa = 4.49DD461 pKa = 3.53TCC463 pKa = 6.15
MM1 pKa = 7.4LSSHH5 pKa = 5.8TFDD8 pKa = 4.7RR9 pKa = 11.84PAAGQTSEE17 pKa = 4.14GVGVVSSEE25 pKa = 4.27VQSQPTGSEE34 pKa = 3.79GFAPHH39 pKa = 6.91TDD41 pKa = 2.87GSQYY45 pKa = 11.31EE46 pKa = 4.37NGEE49 pKa = 4.02TAYY52 pKa = 10.47DD53 pKa = 4.24GSQIEE58 pKa = 4.45SPEE61 pKa = 3.79IPEE64 pKa = 4.5IEE66 pKa = 4.55SNGNGHH72 pKa = 6.38KK73 pKa = 10.23QLLNGGEE80 pKa = 4.19TGFTGVDD87 pKa = 2.9HH88 pKa = 6.9FMAGTSQIQEE98 pKa = 3.92GGGFDD103 pKa = 3.5SFGTSSHH110 pKa = 6.8LEE112 pKa = 3.91TTGVIDD118 pKa = 4.26QSSHH122 pKa = 6.76DD123 pKa = 3.95FLVDD127 pKa = 3.31LMGGIGEE134 pKa = 4.68SYY136 pKa = 11.15GPDD139 pKa = 3.3TQTDD143 pKa = 3.94GLGTLLDD150 pKa = 4.71LLDD153 pKa = 4.68TPPDD157 pKa = 3.67VPPTCLVTDD166 pKa = 4.48AVSTDD171 pKa = 2.89HH172 pKa = 6.74MGLAFQVDD180 pKa = 3.94SAAAGLSPGHH190 pKa = 6.91PSSGGPVLDD199 pKa = 5.32APPDD203 pKa = 4.04SPGLDD208 pKa = 3.3YY209 pKa = 11.6LFVDD213 pKa = 4.78NGNGDD218 pKa = 3.74YY219 pKa = 9.44THH221 pKa = 6.75SVSISDD227 pKa = 4.25NGAQSKK233 pKa = 10.69SPADD237 pKa = 3.69TTDD240 pKa = 2.91SSVVFDD246 pKa = 4.08TMSHH250 pKa = 6.95PDD252 pKa = 3.49HH253 pKa = 7.0FFTDD257 pKa = 4.41YY258 pKa = 11.3SDD260 pKa = 3.4GWVDD264 pKa = 4.17NNGADD269 pKa = 4.47LPEE272 pKa = 4.48TNGNGNGRR280 pKa = 11.84HH281 pKa = 5.55KK282 pKa = 10.62PVVDD286 pKa = 3.74MQKK289 pKa = 10.79GDD291 pKa = 3.69PQGVHH296 pKa = 6.67LDD298 pKa = 3.4ISGTGHH304 pKa = 6.28QDD306 pKa = 2.52AATAMHH312 pKa = 6.39HH313 pKa = 5.46TAVGALDD320 pKa = 5.28LNDD323 pKa = 4.14HH324 pKa = 5.91TLSPYY329 pKa = 10.54TDD331 pKa = 3.15TTGFDD336 pKa = 3.83GVIGAGAQTDD346 pKa = 3.72MAGAGGDD353 pKa = 3.94PATDD357 pKa = 3.68GQTLTDD363 pKa = 3.55MTGQGHH369 pKa = 7.02LAVTDD374 pKa = 3.57GMPNYY379 pKa = 9.25TADD382 pKa = 3.53SVRR385 pKa = 11.84ATGTGFTDD393 pKa = 4.13ASDD396 pKa = 3.61THH398 pKa = 8.17SSMVQTDD405 pKa = 4.1LPVTGDD411 pKa = 3.46PFTGVSSQTDD421 pKa = 3.08AMGTGQPGATEE432 pKa = 4.0QTQTAVSAGEE442 pKa = 4.04QYY444 pKa = 8.74HH445 pKa = 5.93TSGQGFEE452 pKa = 4.14GAEE455 pKa = 3.98NVEE458 pKa = 5.53LEE460 pKa = 4.49DD461 pKa = 3.53TCC463 pKa = 6.15
Molecular weight: 47.41 kDa
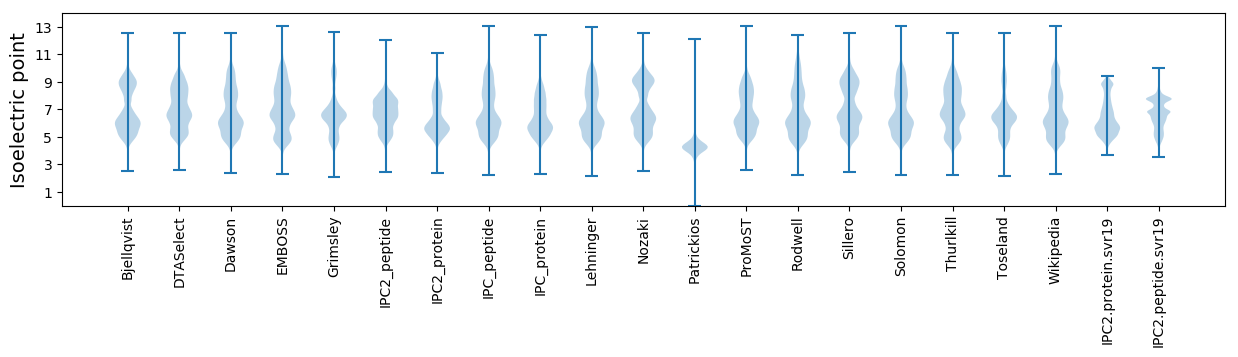
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P6QWA1|A0A6P6QWA1_CARAU LIM/homeobox protein Lhx1 OS=Carassius auratus OX=7957 GN=LOC113114903 PE=4 SV=1
MM1 pKa = 7.76ASHH4 pKa = 5.87KK5 pKa = 8.13TFRR8 pKa = 11.84IKK10 pKa = 10.64RR11 pKa = 11.84FLAKK15 pKa = 9.71KK16 pKa = 9.58QKK18 pKa = 8.69QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84MKK30 pKa = 9.89TGNKK34 pKa = 8.61IRR36 pKa = 11.84YY37 pKa = 7.09NSKK40 pKa = 8.3RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 3.95WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.83LGLL51 pKa = 3.67
MM1 pKa = 7.76ASHH4 pKa = 5.87KK5 pKa = 8.13TFRR8 pKa = 11.84IKK10 pKa = 10.64RR11 pKa = 11.84FLAKK15 pKa = 9.71KK16 pKa = 9.58QKK18 pKa = 8.69QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84MKK30 pKa = 9.89TGNKK34 pKa = 8.61IRR36 pKa = 11.84YY37 pKa = 7.09NSKK40 pKa = 8.3RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 3.95WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.83LGLL51 pKa = 3.67
Molecular weight: 6.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
61822548 |
32 |
30013 |
745.2 |
83.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.212 ± 0.007 | 2.106 ± 0.007 |
5.292 ± 0.006 | 7.291 ± 0.015 |
3.369 ± 0.007 | 6.128 ± 0.014 |
2.595 ± 0.006 | 4.541 ± 0.009 |
5.99 ± 0.018 | 9.045 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.33 ± 0.006 | 3.896 ± 0.006 |
5.847 ± 0.012 | 4.89 ± 0.014 |
5.544 ± 0.008 | 9.124 ± 0.014 |
5.862 ± 0.014 | 6.262 ± 0.014 |
1.066 ± 0.003 | 2.609 ± 0.005 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
