
Xanthomonas virus Cf1c
Taxonomy: Viruses; Monodnaviria; Loebvirae; Hofneiviricota; Faserviricetes; Tubulavirales; Inoviridae; Coriovirus
Average proteome isoelectric point is 8.35
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q38198|Q38198_9VIRU Zot domain-containing protein OS=Xanthomonas virus Cf1c OX=10862 PE=4 SV=1
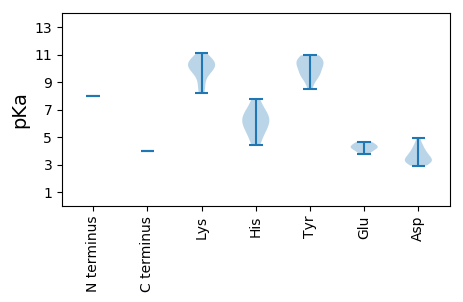
MM1 pKa = 7.97IGDD4 pKa = 3.81TASISLLTGLPGSGKK19 pKa = 8.34SLRR22 pKa = 11.84IIQAIRR28 pKa = 11.84YY29 pKa = 9.0LMDD32 pKa = 4.18KK33 pKa = 9.88GAHH36 pKa = 5.53VYY38 pKa = 9.57VCNIDD43 pKa = 4.48GISVPGTTPWADD55 pKa = 3.15PHH57 pKa = 6.53KK58 pKa = 10.23WQDD61 pKa = 3.48LPAGSILFVDD71 pKa = 4.95EE72 pKa = 4.6AQHH75 pKa = 6.13FFPARR80 pKa = 11.84RR81 pKa = 11.84GGDD84 pKa = 3.26PVEE87 pKa = 4.56TIKK90 pKa = 11.15AMSTIRR96 pKa = 11.84HH97 pKa = 5.97DD98 pKa = 3.9GVRR101 pKa = 11.84LVLATQQPNYY111 pKa = 10.18LDD113 pKa = 3.38TYY115 pKa = 10.94LRR117 pKa = 11.84GLVGYY122 pKa = 9.73HH123 pKa = 5.24EE124 pKa = 4.67HH125 pKa = 7.76LLRR128 pKa = 11.84QSGKK132 pKa = 9.0QKK134 pKa = 9.71TFIFRR139 pKa = 11.84NSQIIEE145 pKa = 4.26EE146 pKa = 4.36VRR148 pKa = 11.84SPLPRR153 pKa = 11.84IKK155 pKa = 10.39KK156 pKa = 9.94LYY158 pKa = 10.38DD159 pKa = 3.24YY160 pKa = 9.6EE161 pKa = 4.33VWKK164 pKa = 10.87QPTEE168 pKa = 3.77CFKK171 pKa = 10.57FYY173 pKa = 10.94KK174 pKa = 9.94SAEE177 pKa = 4.12VHH179 pKa = 4.39TMKK182 pKa = 10.73YY183 pKa = 10.3QMPALVKK190 pKa = 9.54KK191 pKa = 10.45ALMILPVVALLAGGAWYY208 pKa = 9.99AVYY211 pKa = 10.38RR212 pKa = 11.84DD213 pKa = 3.29TMFAKK218 pKa = 10.52KK219 pKa = 10.24ADD221 pKa = 3.69AAPANKK227 pKa = 8.2TAPSGPSLAGTASAGAAARR246 pKa = 11.84PKK248 pKa = 10.47VNSAEE253 pKa = 4.32DD254 pKa = 3.76YY255 pKa = 10.77VGQLVPLVADD265 pKa = 4.41VPWSAPAYY273 pKa = 8.48VDD275 pKa = 3.54RR276 pKa = 11.84PVVSDD281 pKa = 3.01PHH283 pKa = 6.85VYY285 pKa = 10.69CMATEE290 pKa = 4.17NTCRR294 pKa = 11.84CVTEE298 pKa = 3.99QNSRR302 pKa = 11.84VVMRR306 pKa = 11.84DD307 pKa = 3.27DD308 pKa = 3.3VCRR311 pKa = 11.84DD312 pKa = 2.92IARR315 pKa = 11.84WGEE318 pKa = 3.85PYY320 pKa = 10.66NPYY323 pKa = 10.23KK324 pKa = 10.21PPHH327 pKa = 6.52SPAQKK332 pKa = 9.84QQDD335 pKa = 4.03SPVAEE340 pKa = 4.11ATQPKK345 pKa = 8.9PQVPQQSGAVSSSVQRR361 pKa = 11.84ATRR364 pKa = 11.84SLGTFPEE371 pKa = 4.46SPPYY375 pKa = 9.32QTTTYY380 pKa = 9.45TPPTTRR386 pKa = 11.84DD387 pKa = 3.05LL388 pKa = 4.02
MM1 pKa = 7.97IGDD4 pKa = 3.81TASISLLTGLPGSGKK19 pKa = 8.34SLRR22 pKa = 11.84IIQAIRR28 pKa = 11.84YY29 pKa = 9.0LMDD32 pKa = 4.18KK33 pKa = 9.88GAHH36 pKa = 5.53VYY38 pKa = 9.57VCNIDD43 pKa = 4.48GISVPGTTPWADD55 pKa = 3.15PHH57 pKa = 6.53KK58 pKa = 10.23WQDD61 pKa = 3.48LPAGSILFVDD71 pKa = 4.95EE72 pKa = 4.6AQHH75 pKa = 6.13FFPARR80 pKa = 11.84RR81 pKa = 11.84GGDD84 pKa = 3.26PVEE87 pKa = 4.56TIKK90 pKa = 11.15AMSTIRR96 pKa = 11.84HH97 pKa = 5.97DD98 pKa = 3.9GVRR101 pKa = 11.84LVLATQQPNYY111 pKa = 10.18LDD113 pKa = 3.38TYY115 pKa = 10.94LRR117 pKa = 11.84GLVGYY122 pKa = 9.73HH123 pKa = 5.24EE124 pKa = 4.67HH125 pKa = 7.76LLRR128 pKa = 11.84QSGKK132 pKa = 9.0QKK134 pKa = 9.71TFIFRR139 pKa = 11.84NSQIIEE145 pKa = 4.26EE146 pKa = 4.36VRR148 pKa = 11.84SPLPRR153 pKa = 11.84IKK155 pKa = 10.39KK156 pKa = 9.94LYY158 pKa = 10.38DD159 pKa = 3.24YY160 pKa = 9.6EE161 pKa = 4.33VWKK164 pKa = 10.87QPTEE168 pKa = 3.77CFKK171 pKa = 10.57FYY173 pKa = 10.94KK174 pKa = 9.94SAEE177 pKa = 4.12VHH179 pKa = 4.39TMKK182 pKa = 10.73YY183 pKa = 10.3QMPALVKK190 pKa = 9.54KK191 pKa = 10.45ALMILPVVALLAGGAWYY208 pKa = 9.99AVYY211 pKa = 10.38RR212 pKa = 11.84DD213 pKa = 3.29TMFAKK218 pKa = 10.52KK219 pKa = 10.24ADD221 pKa = 3.69AAPANKK227 pKa = 8.2TAPSGPSLAGTASAGAAARR246 pKa = 11.84PKK248 pKa = 10.47VNSAEE253 pKa = 4.32DD254 pKa = 3.76YY255 pKa = 10.77VGQLVPLVADD265 pKa = 4.41VPWSAPAYY273 pKa = 8.48VDD275 pKa = 3.54RR276 pKa = 11.84PVVSDD281 pKa = 3.01PHH283 pKa = 6.85VYY285 pKa = 10.69CMATEE290 pKa = 4.17NTCRR294 pKa = 11.84CVTEE298 pKa = 3.99QNSRR302 pKa = 11.84VVMRR306 pKa = 11.84DD307 pKa = 3.27DD308 pKa = 3.3VCRR311 pKa = 11.84DD312 pKa = 2.92IARR315 pKa = 11.84WGEE318 pKa = 3.85PYY320 pKa = 10.66NPYY323 pKa = 10.23KK324 pKa = 10.21PPHH327 pKa = 6.52SPAQKK332 pKa = 9.84QQDD335 pKa = 4.03SPVAEE340 pKa = 4.11ATQPKK345 pKa = 8.9PQVPQQSGAVSSSVQRR361 pKa = 11.84ATRR364 pKa = 11.84SLGTFPEE371 pKa = 4.46SPPYY375 pKa = 9.32QTTTYY380 pKa = 9.45TPPTTRR386 pKa = 11.84DD387 pKa = 3.05LL388 pKa = 4.02
Molecular weight: 42.78 kDa
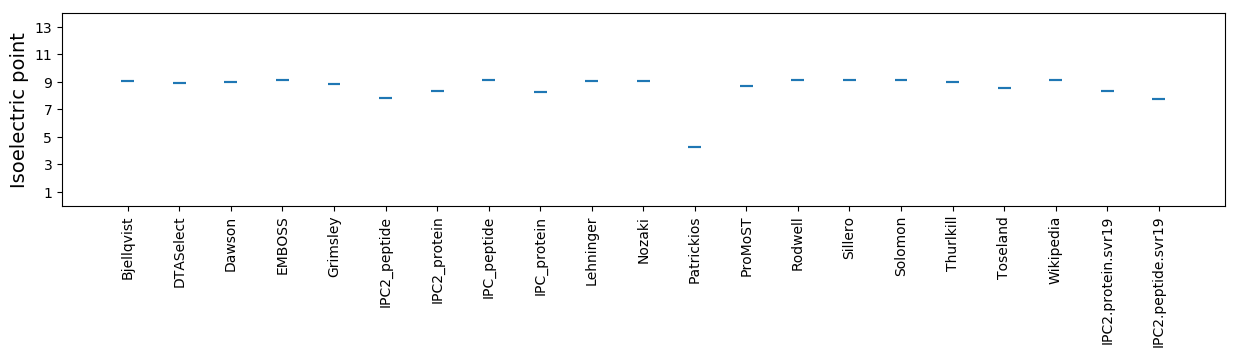
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q38198|Q38198_9VIRU Zot domain-containing protein OS=Xanthomonas virus Cf1c OX=10862 PE=4 SV=1
MM1 pKa = 7.97IGDD4 pKa = 3.81TASISLLTGLPGSGKK19 pKa = 8.34SLRR22 pKa = 11.84IIQAIRR28 pKa = 11.84YY29 pKa = 9.0LMDD32 pKa = 4.18KK33 pKa = 9.88GAHH36 pKa = 5.53VYY38 pKa = 9.57VCNIDD43 pKa = 4.48GISVPGTTPWADD55 pKa = 3.15PHH57 pKa = 6.53KK58 pKa = 10.23WQDD61 pKa = 3.48LPAGSILFVDD71 pKa = 4.95EE72 pKa = 4.6AQHH75 pKa = 6.13FFPARR80 pKa = 11.84RR81 pKa = 11.84GGDD84 pKa = 3.26PVEE87 pKa = 4.56TIKK90 pKa = 11.15AMSTIRR96 pKa = 11.84HH97 pKa = 5.97DD98 pKa = 3.9GVRR101 pKa = 11.84LVLATQQPNYY111 pKa = 10.18LDD113 pKa = 3.38TYY115 pKa = 10.94LRR117 pKa = 11.84GLVGYY122 pKa = 9.73HH123 pKa = 5.24EE124 pKa = 4.67HH125 pKa = 7.76LLRR128 pKa = 11.84QSGKK132 pKa = 9.0QKK134 pKa = 9.71TFIFRR139 pKa = 11.84NSQIIEE145 pKa = 4.26EE146 pKa = 4.36VRR148 pKa = 11.84SPLPRR153 pKa = 11.84IKK155 pKa = 10.39KK156 pKa = 9.94LYY158 pKa = 10.38DD159 pKa = 3.24YY160 pKa = 9.6EE161 pKa = 4.33VWKK164 pKa = 10.87QPTEE168 pKa = 3.77CFKK171 pKa = 10.57FYY173 pKa = 10.94KK174 pKa = 9.94SAEE177 pKa = 4.12VHH179 pKa = 4.39TMKK182 pKa = 10.73YY183 pKa = 10.3QMPALVKK190 pKa = 9.54KK191 pKa = 10.45ALMILPVVALLAGGAWYY208 pKa = 9.99AVYY211 pKa = 10.38RR212 pKa = 11.84DD213 pKa = 3.29TMFAKK218 pKa = 10.52KK219 pKa = 10.24ADD221 pKa = 3.69AAPANKK227 pKa = 8.2TAPSGPSLAGTASAGAAARR246 pKa = 11.84PKK248 pKa = 10.47VNSAEE253 pKa = 4.32DD254 pKa = 3.76YY255 pKa = 10.77VGQLVPLVADD265 pKa = 4.41VPWSAPAYY273 pKa = 8.48VDD275 pKa = 3.54RR276 pKa = 11.84PVVSDD281 pKa = 3.01PHH283 pKa = 6.85VYY285 pKa = 10.69CMATEE290 pKa = 4.17NTCRR294 pKa = 11.84CVTEE298 pKa = 3.99QNSRR302 pKa = 11.84VVMRR306 pKa = 11.84DD307 pKa = 3.27DD308 pKa = 3.3VCRR311 pKa = 11.84DD312 pKa = 2.92IARR315 pKa = 11.84WGEE318 pKa = 3.85PYY320 pKa = 10.66NPYY323 pKa = 10.23KK324 pKa = 10.21PPHH327 pKa = 6.52SPAQKK332 pKa = 9.84QQDD335 pKa = 4.03SPVAEE340 pKa = 4.11ATQPKK345 pKa = 8.9PQVPQQSGAVSSSVQRR361 pKa = 11.84ATRR364 pKa = 11.84SLGTFPEE371 pKa = 4.46SPPYY375 pKa = 9.32QTTTYY380 pKa = 9.45TPPTTRR386 pKa = 11.84DD387 pKa = 3.05LL388 pKa = 4.02
MM1 pKa = 7.97IGDD4 pKa = 3.81TASISLLTGLPGSGKK19 pKa = 8.34SLRR22 pKa = 11.84IIQAIRR28 pKa = 11.84YY29 pKa = 9.0LMDD32 pKa = 4.18KK33 pKa = 9.88GAHH36 pKa = 5.53VYY38 pKa = 9.57VCNIDD43 pKa = 4.48GISVPGTTPWADD55 pKa = 3.15PHH57 pKa = 6.53KK58 pKa = 10.23WQDD61 pKa = 3.48LPAGSILFVDD71 pKa = 4.95EE72 pKa = 4.6AQHH75 pKa = 6.13FFPARR80 pKa = 11.84RR81 pKa = 11.84GGDD84 pKa = 3.26PVEE87 pKa = 4.56TIKK90 pKa = 11.15AMSTIRR96 pKa = 11.84HH97 pKa = 5.97DD98 pKa = 3.9GVRR101 pKa = 11.84LVLATQQPNYY111 pKa = 10.18LDD113 pKa = 3.38TYY115 pKa = 10.94LRR117 pKa = 11.84GLVGYY122 pKa = 9.73HH123 pKa = 5.24EE124 pKa = 4.67HH125 pKa = 7.76LLRR128 pKa = 11.84QSGKK132 pKa = 9.0QKK134 pKa = 9.71TFIFRR139 pKa = 11.84NSQIIEE145 pKa = 4.26EE146 pKa = 4.36VRR148 pKa = 11.84SPLPRR153 pKa = 11.84IKK155 pKa = 10.39KK156 pKa = 9.94LYY158 pKa = 10.38DD159 pKa = 3.24YY160 pKa = 9.6EE161 pKa = 4.33VWKK164 pKa = 10.87QPTEE168 pKa = 3.77CFKK171 pKa = 10.57FYY173 pKa = 10.94KK174 pKa = 9.94SAEE177 pKa = 4.12VHH179 pKa = 4.39TMKK182 pKa = 10.73YY183 pKa = 10.3QMPALVKK190 pKa = 9.54KK191 pKa = 10.45ALMILPVVALLAGGAWYY208 pKa = 9.99AVYY211 pKa = 10.38RR212 pKa = 11.84DD213 pKa = 3.29TMFAKK218 pKa = 10.52KK219 pKa = 10.24ADD221 pKa = 3.69AAPANKK227 pKa = 8.2TAPSGPSLAGTASAGAAARR246 pKa = 11.84PKK248 pKa = 10.47VNSAEE253 pKa = 4.32DD254 pKa = 3.76YY255 pKa = 10.77VGQLVPLVADD265 pKa = 4.41VPWSAPAYY273 pKa = 8.48VDD275 pKa = 3.54RR276 pKa = 11.84PVVSDD281 pKa = 3.01PHH283 pKa = 6.85VYY285 pKa = 10.69CMATEE290 pKa = 4.17NTCRR294 pKa = 11.84CVTEE298 pKa = 3.99QNSRR302 pKa = 11.84VVMRR306 pKa = 11.84DD307 pKa = 3.27DD308 pKa = 3.3VCRR311 pKa = 11.84DD312 pKa = 2.92IARR315 pKa = 11.84WGEE318 pKa = 3.85PYY320 pKa = 10.66NPYY323 pKa = 10.23KK324 pKa = 10.21PPHH327 pKa = 6.52SPAQKK332 pKa = 9.84QQDD335 pKa = 4.03SPVAEE340 pKa = 4.11ATQPKK345 pKa = 8.9PQVPQQSGAVSSSVQRR361 pKa = 11.84ATRR364 pKa = 11.84SLGTFPEE371 pKa = 4.46SPPYY375 pKa = 9.32QTTTYY380 pKa = 9.45TPPTTRR386 pKa = 11.84DD387 pKa = 3.05LL388 pKa = 4.02
Molecular weight: 42.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
388 |
388 |
388 |
388.0 |
42.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.052 ± 0.0 | 1.546 ± 0.0 |
5.412 ± 0.0 | 3.608 ± 0.0 |
2.32 ± 0.0 | 5.928 ± 0.0 |
2.32 ± 0.0 | 4.124 ± 0.0 |
5.412 ± 0.0 | 6.701 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.32 ± 0.0 | 2.062 ± 0.0 |
9.278 ± 0.0 | 5.412 ± 0.0 |
5.67 ± 0.0 | 6.701 ± 0.0 |
6.701 ± 0.0 | 8.247 ± 0.0 |
1.546 ± 0.0 | 4.639 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
