
Sewage-associated gemycircularvirus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemygorvirus; Gemygorvirus sewopo1; Sewage derived gemygorvirus 1
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
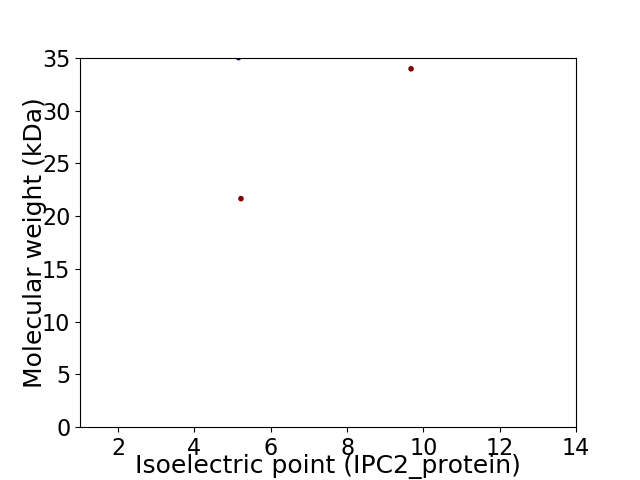
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7CL64|A0A0A7CL64_9VIRU Capsid protein OS=Sewage-associated gemycircularvirus 5 OX=1519400 PE=4 SV=1
MM1 pKa = 7.68AEE3 pKa = 3.77SSFRR7 pKa = 11.84VQSRR11 pKa = 11.84YY12 pKa = 10.07VLLTYY17 pKa = 8.03SQCGEE22 pKa = 4.04LDD24 pKa = 2.95PWAVHH29 pKa = 7.07DD30 pKa = 5.8SITSYY35 pKa = 10.23PAEE38 pKa = 3.97CLIGRR43 pKa = 11.84EE44 pKa = 3.92NHH46 pKa = 6.51ADD48 pKa = 3.55GGIHH52 pKa = 6.42LHH54 pKa = 6.48AFVDD58 pKa = 4.58FGRR61 pKa = 11.84RR62 pKa = 11.84INLRR66 pKa = 11.84DD67 pKa = 3.19SRR69 pKa = 11.84RR70 pKa = 11.84FDD72 pKa = 3.29VEE74 pKa = 4.77GYY76 pKa = 9.96HH77 pKa = 7.3PNIQPCGRR85 pKa = 11.84TPQKK89 pKa = 10.33MLDD92 pKa = 3.61YY93 pKa = 10.33AIKK96 pKa = 10.91DD97 pKa = 3.49GDD99 pKa = 4.03VVAGGLSPTVGTEE112 pKa = 3.71VQGADD117 pKa = 3.52NVWARR122 pKa = 11.84IADD125 pKa = 3.77AASVEE130 pKa = 4.52EE131 pKa = 4.31FWEE134 pKa = 4.38LARR137 pKa = 11.84EE138 pKa = 4.16LAPRR142 pKa = 11.84ALLCNHH148 pKa = 5.94QSLRR152 pKa = 11.84AYY154 pKa = 10.25AEE156 pKa = 3.52WHH158 pKa = 5.67YY159 pKa = 11.35RR160 pKa = 11.84PATVAYY166 pKa = 6.72EE167 pKa = 4.18HH168 pKa = 7.23PPGLHH173 pKa = 6.47LCTGGVPEE181 pKa = 4.5LDD183 pKa = 2.61AWVRR187 pKa = 11.84DD188 pKa = 3.56NLLILWGEE196 pKa = 4.02TRR198 pKa = 11.84LGKK201 pKa = 7.89TVWARR206 pKa = 11.84SLGPHH211 pKa = 6.51VYY213 pKa = 10.27CCLQWNVDD221 pKa = 3.76DD222 pKa = 5.57LKK224 pKa = 11.47AGLEE228 pKa = 4.03DD229 pKa = 3.24AKK231 pKa = 11.24YY232 pKa = 10.74AVLDD236 pKa = 4.77DD237 pKa = 3.68IQGNFQFFPAYY248 pKa = 9.6KK249 pKa = 9.96GWLGGQATFTVTDD262 pKa = 4.1KK263 pKa = 11.38YY264 pKa = 10.68RR265 pKa = 11.84GKK267 pKa = 7.72TTVQWGRR274 pKa = 11.84PTIWLMNDD282 pKa = 3.12DD283 pKa = 4.37PEE285 pKa = 4.6EE286 pKa = 4.26VGHH289 pKa = 7.26VDD291 pKa = 5.07LNWLRR296 pKa = 11.84GNCDD300 pKa = 3.38IIHH303 pKa = 6.0LTEE306 pKa = 4.29SLISSS311 pKa = 4.28
MM1 pKa = 7.68AEE3 pKa = 3.77SSFRR7 pKa = 11.84VQSRR11 pKa = 11.84YY12 pKa = 10.07VLLTYY17 pKa = 8.03SQCGEE22 pKa = 4.04LDD24 pKa = 2.95PWAVHH29 pKa = 7.07DD30 pKa = 5.8SITSYY35 pKa = 10.23PAEE38 pKa = 3.97CLIGRR43 pKa = 11.84EE44 pKa = 3.92NHH46 pKa = 6.51ADD48 pKa = 3.55GGIHH52 pKa = 6.42LHH54 pKa = 6.48AFVDD58 pKa = 4.58FGRR61 pKa = 11.84RR62 pKa = 11.84INLRR66 pKa = 11.84DD67 pKa = 3.19SRR69 pKa = 11.84RR70 pKa = 11.84FDD72 pKa = 3.29VEE74 pKa = 4.77GYY76 pKa = 9.96HH77 pKa = 7.3PNIQPCGRR85 pKa = 11.84TPQKK89 pKa = 10.33MLDD92 pKa = 3.61YY93 pKa = 10.33AIKK96 pKa = 10.91DD97 pKa = 3.49GDD99 pKa = 4.03VVAGGLSPTVGTEE112 pKa = 3.71VQGADD117 pKa = 3.52NVWARR122 pKa = 11.84IADD125 pKa = 3.77AASVEE130 pKa = 4.52EE131 pKa = 4.31FWEE134 pKa = 4.38LARR137 pKa = 11.84EE138 pKa = 4.16LAPRR142 pKa = 11.84ALLCNHH148 pKa = 5.94QSLRR152 pKa = 11.84AYY154 pKa = 10.25AEE156 pKa = 3.52WHH158 pKa = 5.67YY159 pKa = 11.35RR160 pKa = 11.84PATVAYY166 pKa = 6.72EE167 pKa = 4.18HH168 pKa = 7.23PPGLHH173 pKa = 6.47LCTGGVPEE181 pKa = 4.5LDD183 pKa = 2.61AWVRR187 pKa = 11.84DD188 pKa = 3.56NLLILWGEE196 pKa = 4.02TRR198 pKa = 11.84LGKK201 pKa = 7.89TVWARR206 pKa = 11.84SLGPHH211 pKa = 6.51VYY213 pKa = 10.27CCLQWNVDD221 pKa = 3.76DD222 pKa = 5.57LKK224 pKa = 11.47AGLEE228 pKa = 4.03DD229 pKa = 3.24AKK231 pKa = 11.24YY232 pKa = 10.74AVLDD236 pKa = 4.77DD237 pKa = 3.68IQGNFQFFPAYY248 pKa = 9.6KK249 pKa = 9.96GWLGGQATFTVTDD262 pKa = 4.1KK263 pKa = 11.38YY264 pKa = 10.68RR265 pKa = 11.84GKK267 pKa = 7.72TTVQWGRR274 pKa = 11.84PTIWLMNDD282 pKa = 3.12DD283 pKa = 4.37PEE285 pKa = 4.6EE286 pKa = 4.26VGHH289 pKa = 7.26VDD291 pKa = 5.07LNWLRR296 pKa = 11.84GNCDD300 pKa = 3.38IIHH303 pKa = 6.0LTEE306 pKa = 4.29SLISSS311 pKa = 4.28
Molecular weight: 35.03 kDa
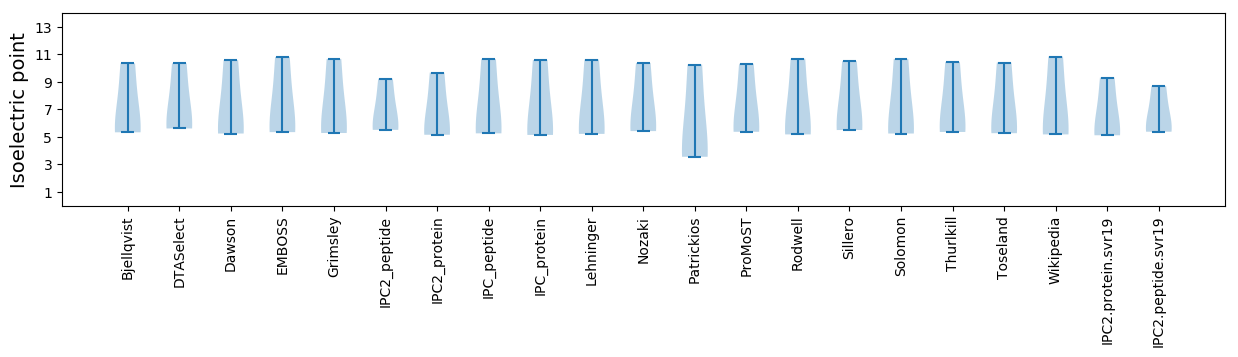
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7CL64|A0A0A7CL64_9VIRU Capsid protein OS=Sewage-associated gemycircularvirus 5 OX=1519400 PE=4 SV=1
MM1 pKa = 7.4PRR3 pKa = 11.84RR4 pKa = 11.84YY5 pKa = 7.91TRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84ARR13 pKa = 11.84TTRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84PAFRR22 pKa = 11.84KK23 pKa = 8.44ATTTRR28 pKa = 11.84GRR30 pKa = 11.84RR31 pKa = 11.84TRR33 pKa = 11.84SFARR37 pKa = 11.84TRR39 pKa = 11.84HH40 pKa = 4.01SRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84ILNITSIKK52 pKa = 10.24KK53 pKa = 9.42HH54 pKa = 6.76DD55 pKa = 3.97NMLPAVRR62 pKa = 11.84TADD65 pKa = 2.95GGYY68 pKa = 9.26SQNPLTTTTGFTSLFMPTARR88 pKa = 11.84EE89 pKa = 3.86LAHH92 pKa = 5.68TAEE95 pKa = 4.92GEE97 pKa = 4.42STRR100 pKa = 11.84QRR102 pKa = 11.84QTTFSVGYY110 pKa = 9.58KK111 pKa = 9.78EE112 pKa = 4.63RR113 pKa = 11.84LQIDD117 pKa = 3.92VQGGGVWKK125 pKa = 9.58WRR127 pKa = 11.84RR128 pKa = 11.84LIFTYY133 pKa = 10.33KK134 pKa = 10.42GDD136 pKa = 3.85SLFDD140 pKa = 4.33GDD142 pKa = 4.59PTWTQPFWDD151 pKa = 4.78KK152 pKa = 10.94STDD155 pKa = 3.53PDD157 pKa = 3.8GVDD160 pKa = 3.32MVRR163 pKa = 11.84LIAQPTLDD171 pKa = 3.65QHH173 pKa = 6.57ALWRR177 pKa = 11.84RR178 pKa = 11.84ILWDD182 pKa = 3.48GEE184 pKa = 4.29EE185 pKa = 4.33GVDD188 pKa = 3.36WNSEE192 pKa = 3.7FTAKK196 pKa = 10.26VDD198 pKa = 3.51TSRR201 pKa = 11.84ITPRR205 pKa = 11.84YY206 pKa = 10.0DD207 pKa = 3.0RR208 pKa = 11.84TITLNPGNEE217 pKa = 4.22SGMSRR222 pKa = 11.84TFRR225 pKa = 11.84LWHH228 pKa = 5.54PTRR231 pKa = 11.84KK232 pKa = 10.33NIVYY236 pKa = 10.46DD237 pKa = 3.82DD238 pKa = 4.79DD239 pKa = 4.43EE240 pKa = 5.93FGGAPNGGGAYY251 pKa = 9.46TSVTGKK257 pKa = 9.83PGMGDD262 pKa = 3.73LYY264 pKa = 11.33VYY266 pKa = 10.13DD267 pKa = 3.37IVYY270 pKa = 10.54NAVPAAAGNQALVFNPEE287 pKa = 3.3GTYY290 pKa = 10.18YY291 pKa = 9.06WHH293 pKa = 6.98EE294 pKa = 4.02RR295 pKa = 3.5
MM1 pKa = 7.4PRR3 pKa = 11.84RR4 pKa = 11.84YY5 pKa = 7.91TRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84ARR13 pKa = 11.84TTRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84PAFRR22 pKa = 11.84KK23 pKa = 8.44ATTTRR28 pKa = 11.84GRR30 pKa = 11.84RR31 pKa = 11.84TRR33 pKa = 11.84SFARR37 pKa = 11.84TRR39 pKa = 11.84HH40 pKa = 4.01SRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84ILNITSIKK52 pKa = 10.24KK53 pKa = 9.42HH54 pKa = 6.76DD55 pKa = 3.97NMLPAVRR62 pKa = 11.84TADD65 pKa = 2.95GGYY68 pKa = 9.26SQNPLTTTTGFTSLFMPTARR88 pKa = 11.84EE89 pKa = 3.86LAHH92 pKa = 5.68TAEE95 pKa = 4.92GEE97 pKa = 4.42STRR100 pKa = 11.84QRR102 pKa = 11.84QTTFSVGYY110 pKa = 9.58KK111 pKa = 9.78EE112 pKa = 4.63RR113 pKa = 11.84LQIDD117 pKa = 3.92VQGGGVWKK125 pKa = 9.58WRR127 pKa = 11.84RR128 pKa = 11.84LIFTYY133 pKa = 10.33KK134 pKa = 10.42GDD136 pKa = 3.85SLFDD140 pKa = 4.33GDD142 pKa = 4.59PTWTQPFWDD151 pKa = 4.78KK152 pKa = 10.94STDD155 pKa = 3.53PDD157 pKa = 3.8GVDD160 pKa = 3.32MVRR163 pKa = 11.84LIAQPTLDD171 pKa = 3.65QHH173 pKa = 6.57ALWRR177 pKa = 11.84RR178 pKa = 11.84ILWDD182 pKa = 3.48GEE184 pKa = 4.29EE185 pKa = 4.33GVDD188 pKa = 3.36WNSEE192 pKa = 3.7FTAKK196 pKa = 10.26VDD198 pKa = 3.51TSRR201 pKa = 11.84ITPRR205 pKa = 11.84YY206 pKa = 10.0DD207 pKa = 3.0RR208 pKa = 11.84TITLNPGNEE217 pKa = 4.22SGMSRR222 pKa = 11.84TFRR225 pKa = 11.84LWHH228 pKa = 5.54PTRR231 pKa = 11.84KK232 pKa = 10.33NIVYY236 pKa = 10.46DD237 pKa = 3.82DD238 pKa = 4.79DD239 pKa = 4.43EE240 pKa = 5.93FGGAPNGGGAYY251 pKa = 9.46TSVTGKK257 pKa = 9.83PGMGDD262 pKa = 3.73LYY264 pKa = 11.33VYY266 pKa = 10.13DD267 pKa = 3.37IVYY270 pKa = 10.54NAVPAAAGNQALVFNPEE287 pKa = 3.3GTYY290 pKa = 10.18YY291 pKa = 9.06WHH293 pKa = 6.98EE294 pKa = 4.02RR295 pKa = 3.5
Molecular weight: 33.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
801 |
195 |
311 |
267.0 |
30.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.115 ± 0.839 | 1.623 ± 0.783 |
6.991 ± 0.199 | 5.243 ± 0.722 |
3.246 ± 0.391 | 8.739 ± 0.038 |
3.371 ± 0.637 | 3.87 ± 0.152 |
2.497 ± 0.495 | 7.99 ± 1.308 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.373 ± 0.321 | 3.496 ± 0.136 |
5.243 ± 0.207 | 3.246 ± 0.147 |
9.114 ± 1.88 | 4.869 ± 0.239 |
7.491 ± 2.093 | 6.242 ± 0.718 |
3.246 ± 0.305 | 3.995 ± 0.067 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
