
Okra leaf curl virus-[Cameroon]
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus; unclassified Begomovirus; Okra leaf curl virus
Average proteome isoelectric point is 7.42
Get precalculated fractions of proteins
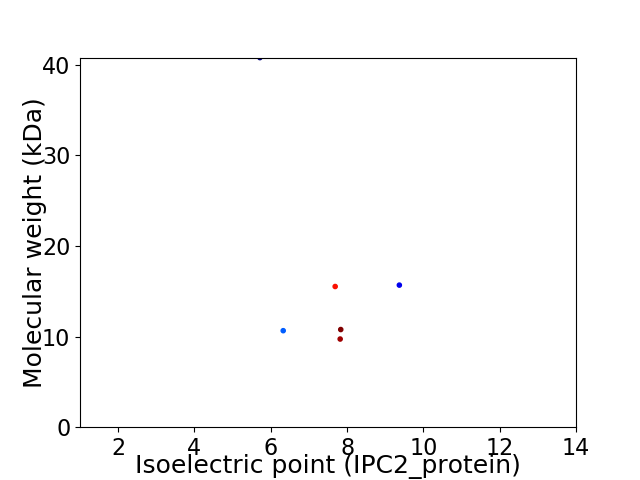
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|C5J4Q8|C5J4Q8_9GEMI Uncharacterized protein OS=Okra leaf curl virus-[Cameroon] OX=627503 GN=C4 PE=3 SV=1
MM1 pKa = 7.64APNKK5 pKa = 10.05RR6 pKa = 11.84FQIYY10 pKa = 10.34AKK12 pKa = 10.52NFFLTFPKK20 pKa = 10.45CSLSKK25 pKa = 11.15EE26 pKa = 4.04EE27 pKa = 5.44ALEE30 pKa = 3.78QLQQISTASNKK41 pKa = 10.08KK42 pKa = 9.09YY43 pKa = 10.71IKK45 pKa = 9.68ICRR48 pKa = 11.84EE49 pKa = 3.81LHH51 pKa = 6.97EE52 pKa = 5.88DD53 pKa = 3.95GQPHH57 pKa = 6.66LHH59 pKa = 6.38VLLQFEE65 pKa = 5.47GKK67 pKa = 10.02FKK69 pKa = 10.87CQNQRR74 pKa = 11.84LFDD77 pKa = 4.01LVSPNRR83 pKa = 11.84SAHH86 pKa = 5.06FHH88 pKa = 6.56PNIQGAKK95 pKa = 9.25SSSDD99 pKa = 3.11VKK101 pKa = 11.24SYY103 pKa = 10.79IDD105 pKa = 3.7KK106 pKa = 11.33DD107 pKa = 3.49GDD109 pKa = 3.67TLEE112 pKa = 4.14WGEE115 pKa = 3.84FQIDD119 pKa = 3.21GRR121 pKa = 11.84SARR124 pKa = 11.84GGQQTANDD132 pKa = 4.05AYY134 pKa = 10.69AAALNAGSKK143 pKa = 9.22TEE145 pKa = 3.75ALRR148 pKa = 11.84VIRR151 pKa = 11.84EE152 pKa = 4.09LAPKK156 pKa = 10.51DD157 pKa = 4.02FVLQYY162 pKa = 11.26HH163 pKa = 5.78NLINNLDD170 pKa = 4.39RR171 pKa = 11.84IFQEE175 pKa = 4.24PPAPYY180 pKa = 9.99VSPFLSSSFDD190 pKa = 3.44QVPEE194 pKa = 3.98EE195 pKa = 4.13LEE197 pKa = 3.58EE198 pKa = 3.9WAAEE202 pKa = 4.15NVVEE206 pKa = 4.32AAARR210 pKa = 11.84PGRR213 pKa = 11.84PISIVIEE220 pKa = 4.35GEE222 pKa = 3.93SRR224 pKa = 11.84TGKK227 pKa = 7.7TVWARR232 pKa = 11.84SLGPHH237 pKa = 6.71NYY239 pKa = 10.18LCGHH243 pKa = 7.38LDD245 pKa = 3.88LSPKK249 pKa = 10.04VFSNDD254 pKa = 1.85AWYY257 pKa = 10.93NVIDD261 pKa = 5.2DD262 pKa = 4.41VDD264 pKa = 3.61PHH266 pKa = 5.85YY267 pKa = 11.0LKK269 pKa = 10.7HH270 pKa = 6.2FKK272 pKa = 10.76EE273 pKa = 4.49FMGAQKK279 pKa = 10.58DD280 pKa = 3.71WQSNTKK286 pKa = 9.54YY287 pKa = 10.57GKK289 pKa = 9.12PVQIKK294 pKa = 10.41GGIPTIFLCNPGPNSSYY311 pKa = 11.09KK312 pKa = 10.65DD313 pKa = 3.59YY314 pKa = 11.45LDD316 pKa = 3.85EE317 pKa = 4.94EE318 pKa = 4.85KK319 pKa = 10.74NAHH322 pKa = 5.77LKK324 pKa = 9.97SWALKK329 pKa = 8.81NATFITLSYY338 pKa = 10.0PLYY341 pKa = 10.72SGTNQSPASGGQEE354 pKa = 3.63EE355 pKa = 5.28SNQEE359 pKa = 3.99TQDD362 pKa = 3.25
MM1 pKa = 7.64APNKK5 pKa = 10.05RR6 pKa = 11.84FQIYY10 pKa = 10.34AKK12 pKa = 10.52NFFLTFPKK20 pKa = 10.45CSLSKK25 pKa = 11.15EE26 pKa = 4.04EE27 pKa = 5.44ALEE30 pKa = 3.78QLQQISTASNKK41 pKa = 10.08KK42 pKa = 9.09YY43 pKa = 10.71IKK45 pKa = 9.68ICRR48 pKa = 11.84EE49 pKa = 3.81LHH51 pKa = 6.97EE52 pKa = 5.88DD53 pKa = 3.95GQPHH57 pKa = 6.66LHH59 pKa = 6.38VLLQFEE65 pKa = 5.47GKK67 pKa = 10.02FKK69 pKa = 10.87CQNQRR74 pKa = 11.84LFDD77 pKa = 4.01LVSPNRR83 pKa = 11.84SAHH86 pKa = 5.06FHH88 pKa = 6.56PNIQGAKK95 pKa = 9.25SSSDD99 pKa = 3.11VKK101 pKa = 11.24SYY103 pKa = 10.79IDD105 pKa = 3.7KK106 pKa = 11.33DD107 pKa = 3.49GDD109 pKa = 3.67TLEE112 pKa = 4.14WGEE115 pKa = 3.84FQIDD119 pKa = 3.21GRR121 pKa = 11.84SARR124 pKa = 11.84GGQQTANDD132 pKa = 4.05AYY134 pKa = 10.69AAALNAGSKK143 pKa = 9.22TEE145 pKa = 3.75ALRR148 pKa = 11.84VIRR151 pKa = 11.84EE152 pKa = 4.09LAPKK156 pKa = 10.51DD157 pKa = 4.02FVLQYY162 pKa = 11.26HH163 pKa = 5.78NLINNLDD170 pKa = 4.39RR171 pKa = 11.84IFQEE175 pKa = 4.24PPAPYY180 pKa = 9.99VSPFLSSSFDD190 pKa = 3.44QVPEE194 pKa = 3.98EE195 pKa = 4.13LEE197 pKa = 3.58EE198 pKa = 3.9WAAEE202 pKa = 4.15NVVEE206 pKa = 4.32AAARR210 pKa = 11.84PGRR213 pKa = 11.84PISIVIEE220 pKa = 4.35GEE222 pKa = 3.93SRR224 pKa = 11.84TGKK227 pKa = 7.7TVWARR232 pKa = 11.84SLGPHH237 pKa = 6.71NYY239 pKa = 10.18LCGHH243 pKa = 7.38LDD245 pKa = 3.88LSPKK249 pKa = 10.04VFSNDD254 pKa = 1.85AWYY257 pKa = 10.93NVIDD261 pKa = 5.2DD262 pKa = 4.41VDD264 pKa = 3.61PHH266 pKa = 5.85YY267 pKa = 11.0LKK269 pKa = 10.7HH270 pKa = 6.2FKK272 pKa = 10.76EE273 pKa = 4.49FMGAQKK279 pKa = 10.58DD280 pKa = 3.71WQSNTKK286 pKa = 9.54YY287 pKa = 10.57GKK289 pKa = 9.12PVQIKK294 pKa = 10.41GGIPTIFLCNPGPNSSYY311 pKa = 11.09KK312 pKa = 10.65DD313 pKa = 3.59YY314 pKa = 11.45LDD316 pKa = 3.85EE317 pKa = 4.94EE318 pKa = 4.85KK319 pKa = 10.74NAHH322 pKa = 5.77LKK324 pKa = 9.97SWALKK329 pKa = 8.81NATFITLSYY338 pKa = 10.0PLYY341 pKa = 10.72SGTNQSPASGGQEE354 pKa = 3.63EE355 pKa = 5.28SNQEE359 pKa = 3.99TQDD362 pKa = 3.25
Molecular weight: 40.8 kDa
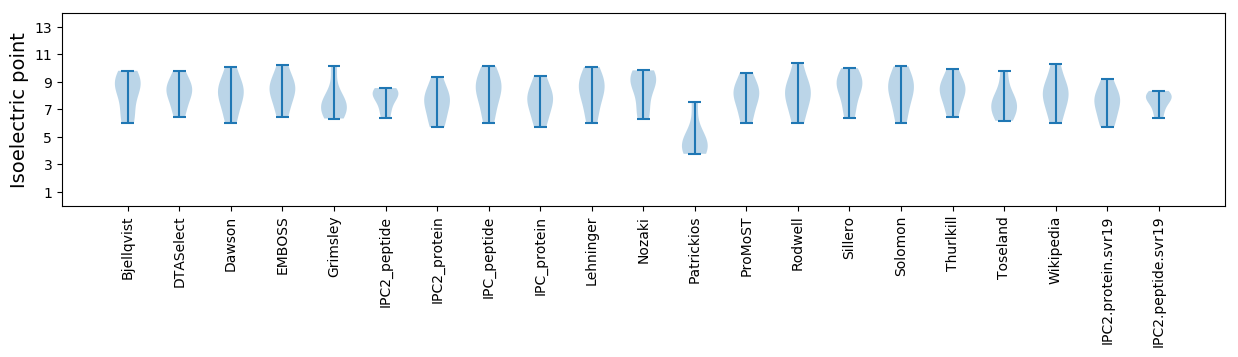
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C5J4Q6|C5J4Q6_9GEMI Transcriptional activator protein OS=Okra leaf curl virus-[Cameroon] OX=627503 PE=3 SV=1
MM1 pKa = 8.27DD2 pKa = 4.36SRR4 pKa = 11.84TGEE7 pKa = 5.1LITAHH12 pKa = 4.42QTEE15 pKa = 4.56NGVLIWTINNPLYY28 pKa = 10.71FKK30 pKa = 9.77TIKK33 pKa = 10.11EE34 pKa = 3.98IPLTHH39 pKa = 6.84GNQTMVEE46 pKa = 3.86MQIRR50 pKa = 11.84FNYY53 pKa = 9.94NLRR56 pKa = 11.84KK57 pKa = 9.59EE58 pKa = 4.28LGIHH62 pKa = 5.36KK63 pKa = 9.9CFMNFRR69 pKa = 11.84VWTISRR75 pKa = 11.84PPTGLFLNVFRR86 pKa = 11.84KK87 pKa = 10.15QIMKK91 pKa = 9.64YY92 pKa = 10.0LYY94 pKa = 8.72RR95 pKa = 11.84LGVISINNVIRR106 pKa = 11.84AVNHH110 pKa = 5.07VLYY113 pKa = 10.55DD114 pKa = 3.56VLQTTVASEE123 pKa = 4.24FTHH126 pKa = 6.83NIQIKK131 pKa = 10.32LYY133 pKa = 10.05
MM1 pKa = 8.27DD2 pKa = 4.36SRR4 pKa = 11.84TGEE7 pKa = 5.1LITAHH12 pKa = 4.42QTEE15 pKa = 4.56NGVLIWTINNPLYY28 pKa = 10.71FKK30 pKa = 9.77TIKK33 pKa = 10.11EE34 pKa = 3.98IPLTHH39 pKa = 6.84GNQTMVEE46 pKa = 3.86MQIRR50 pKa = 11.84FNYY53 pKa = 9.94NLRR56 pKa = 11.84KK57 pKa = 9.59EE58 pKa = 4.28LGIHH62 pKa = 5.36KK63 pKa = 9.9CFMNFRR69 pKa = 11.84VWTISRR75 pKa = 11.84PPTGLFLNVFRR86 pKa = 11.84KK87 pKa = 10.15QIMKK91 pKa = 9.64YY92 pKa = 10.0LYY94 pKa = 8.72RR95 pKa = 11.84LGVISINNVIRR106 pKa = 11.84AVNHH110 pKa = 5.07VLYY113 pKa = 10.55DD114 pKa = 3.56VLQTTVASEE123 pKa = 4.24FTHH126 pKa = 6.83NIQIKK131 pKa = 10.32LYY133 pKa = 10.05
Molecular weight: 15.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
902 |
85 |
362 |
150.3 |
17.2 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.208 ± 1.109 | 2.106 ± 0.534 |
4.878 ± 0.98 | 5.765 ± 0.652 |
4.213 ± 0.536 | 5.1 ± 0.588 |
3.659 ± 0.494 | 6.208 ± 0.983 |
5.322 ± 0.953 | 8.093 ± 0.921 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.328 ± 0.849 | 5.987 ± 0.643 |
5.322 ± 0.588 | 5.654 ± 0.725 |
6.319 ± 1.455 | 7.539 ± 1.402 |
5.765 ± 1.256 | 4.545 ± 0.774 |
1.33 ± 0.224 | 3.659 ± 0.638 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
