
Canine feces-associated gemycircularvirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykibivirus; Gemykibivirus canfam1
Average proteome isoelectric point is 7.34
Get precalculated fractions of proteins
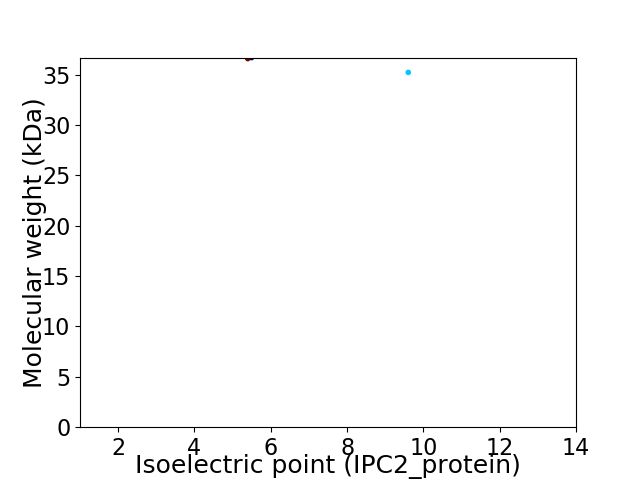
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A221LEC1|A0A221LEC1_9VIRU Replication-associated protein OS=Canine feces-associated gemycircularvirus OX=2017709 PE=3 SV=1
MM1 pKa = 8.01PFRR4 pKa = 11.84FQAKK8 pKa = 9.66YY9 pKa = 11.01GLLTYY14 pKa = 7.94SQCGDD19 pKa = 3.94LDD21 pKa = 3.95PFSVVDD27 pKa = 3.46MLGDD31 pKa = 4.02LGAEE35 pKa = 4.07CIVGRR40 pKa = 11.84EE41 pKa = 3.95GHH43 pKa = 6.0TDD45 pKa = 3.22GGIHH49 pKa = 6.2LHH51 pKa = 6.04AFFMFEE57 pKa = 4.2RR58 pKa = 11.84KK59 pKa = 9.3FSSRR63 pKa = 11.84DD64 pKa = 3.14VRR66 pKa = 11.84VFDD69 pKa = 5.24VDD71 pKa = 3.67GQHH74 pKa = 6.73PNVRR78 pKa = 11.84SGWSTPEE85 pKa = 4.11KK86 pKa = 10.07GWDD89 pKa = 3.28YY90 pKa = 9.74ATKK93 pKa = 10.69YY94 pKa = 10.38GDD96 pKa = 3.57VVAGGLEE103 pKa = 4.04RR104 pKa = 11.84PGNRR108 pKa = 11.84VSASSSVWATILLSEE123 pKa = 4.41DD124 pKa = 3.29RR125 pKa = 11.84EE126 pKa = 4.89SFFEE130 pKa = 4.08TCEE133 pKa = 3.72RR134 pKa = 11.84LAPGTLLRR142 pKa = 11.84NFTSLRR148 pKa = 11.84CYY150 pKa = 10.73ADD152 pKa = 2.62WRR154 pKa = 11.84YY155 pKa = 9.96RR156 pKa = 11.84PEE158 pKa = 4.35PEE160 pKa = 4.67PYY162 pKa = 8.92IHH164 pKa = 7.1PPIISFEE171 pKa = 4.2TAGFPEE177 pKa = 4.96LDD179 pKa = 2.97QWVQQNLVGPRR190 pKa = 11.84VEE192 pKa = 4.43GNIGRR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84SLVLYY204 pKa = 10.49GEE206 pKa = 4.33SRR208 pKa = 11.84TGKK211 pKa = 7.35TLWARR216 pKa = 11.84SLGHH220 pKa = 6.17HH221 pKa = 7.12AYY223 pKa = 10.52FGGLFSLDD231 pKa = 3.58EE232 pKa = 4.44SVDD235 pKa = 3.77DD236 pKa = 3.42VDD238 pKa = 4.83YY239 pKa = 11.54AIFDD243 pKa = 4.01DD244 pKa = 4.22MQGGLSFFHH253 pKa = 7.23SYY255 pKa = 10.69KK256 pKa = 10.47FWLGCQLEE264 pKa = 4.98FWATDD269 pKa = 3.37KK270 pKa = 11.1YY271 pKa = 10.95RR272 pKa = 11.84HH273 pKa = 5.25KK274 pKa = 11.0QKK276 pKa = 10.54IKK278 pKa = 9.12WGRR281 pKa = 11.84PCIYY285 pKa = 10.02ISNNNPLADD294 pKa = 3.92AGVDD298 pKa = 3.86VDD300 pKa = 4.63WLMINCTFVCIDD312 pKa = 3.41QPIFRR317 pKa = 11.84ANTAA321 pKa = 3.21
MM1 pKa = 8.01PFRR4 pKa = 11.84FQAKK8 pKa = 9.66YY9 pKa = 11.01GLLTYY14 pKa = 7.94SQCGDD19 pKa = 3.94LDD21 pKa = 3.95PFSVVDD27 pKa = 3.46MLGDD31 pKa = 4.02LGAEE35 pKa = 4.07CIVGRR40 pKa = 11.84EE41 pKa = 3.95GHH43 pKa = 6.0TDD45 pKa = 3.22GGIHH49 pKa = 6.2LHH51 pKa = 6.04AFFMFEE57 pKa = 4.2RR58 pKa = 11.84KK59 pKa = 9.3FSSRR63 pKa = 11.84DD64 pKa = 3.14VRR66 pKa = 11.84VFDD69 pKa = 5.24VDD71 pKa = 3.67GQHH74 pKa = 6.73PNVRR78 pKa = 11.84SGWSTPEE85 pKa = 4.11KK86 pKa = 10.07GWDD89 pKa = 3.28YY90 pKa = 9.74ATKK93 pKa = 10.69YY94 pKa = 10.38GDD96 pKa = 3.57VVAGGLEE103 pKa = 4.04RR104 pKa = 11.84PGNRR108 pKa = 11.84VSASSSVWATILLSEE123 pKa = 4.41DD124 pKa = 3.29RR125 pKa = 11.84EE126 pKa = 4.89SFFEE130 pKa = 4.08TCEE133 pKa = 3.72RR134 pKa = 11.84LAPGTLLRR142 pKa = 11.84NFTSLRR148 pKa = 11.84CYY150 pKa = 10.73ADD152 pKa = 2.62WRR154 pKa = 11.84YY155 pKa = 9.96RR156 pKa = 11.84PEE158 pKa = 4.35PEE160 pKa = 4.67PYY162 pKa = 8.92IHH164 pKa = 7.1PPIISFEE171 pKa = 4.2TAGFPEE177 pKa = 4.96LDD179 pKa = 2.97QWVQQNLVGPRR190 pKa = 11.84VEE192 pKa = 4.43GNIGRR197 pKa = 11.84RR198 pKa = 11.84RR199 pKa = 11.84SLVLYY204 pKa = 10.49GEE206 pKa = 4.33SRR208 pKa = 11.84TGKK211 pKa = 7.35TLWARR216 pKa = 11.84SLGHH220 pKa = 6.17HH221 pKa = 7.12AYY223 pKa = 10.52FGGLFSLDD231 pKa = 3.58EE232 pKa = 4.44SVDD235 pKa = 3.77DD236 pKa = 3.42VDD238 pKa = 4.83YY239 pKa = 11.54AIFDD243 pKa = 4.01DD244 pKa = 4.22MQGGLSFFHH253 pKa = 7.23SYY255 pKa = 10.69KK256 pKa = 10.47FWLGCQLEE264 pKa = 4.98FWATDD269 pKa = 3.37KK270 pKa = 11.1YY271 pKa = 10.95RR272 pKa = 11.84HH273 pKa = 5.25KK274 pKa = 11.0QKK276 pKa = 10.54IKK278 pKa = 9.12WGRR281 pKa = 11.84PCIYY285 pKa = 10.02ISNNNPLADD294 pKa = 3.92AGVDD298 pKa = 3.86VDD300 pKa = 4.63WLMINCTFVCIDD312 pKa = 3.41QPIFRR317 pKa = 11.84ANTAA321 pKa = 3.21
Molecular weight: 36.59 kDa
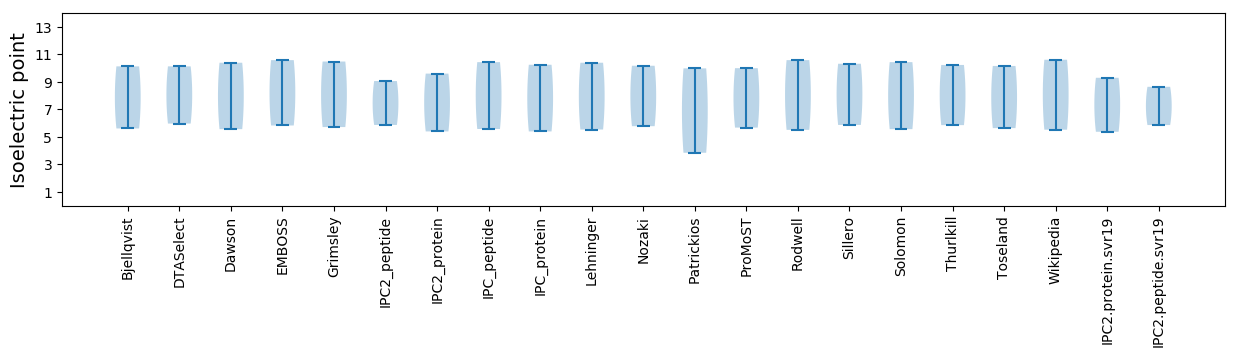
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A221LEC1|A0A221LEC1_9VIRU Replication-associated protein OS=Canine feces-associated gemycircularvirus OX=2017709 PE=3 SV=1
MM1 pKa = 7.54AYY3 pKa = 10.05SRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 8.43SAPRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84FRR16 pKa = 11.84RR17 pKa = 11.84GFKK20 pKa = 10.05RR21 pKa = 11.84RR22 pKa = 11.84SSRR25 pKa = 11.84LAYY28 pKa = 9.67RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84PYY34 pKa = 10.04VRR36 pKa = 11.84RR37 pKa = 11.84PYY39 pKa = 10.54RR40 pKa = 11.84KK41 pKa = 8.49RR42 pKa = 11.84TWTTRR47 pKa = 11.84AILNKK52 pKa = 9.05TSKK55 pKa = 10.61KK56 pKa = 9.44KK57 pKa = 9.17TDD59 pKa = 3.39TMQVWSDD66 pKa = 3.3TQTPGTYY73 pKa = 9.91VPGGAVIPAGTGAVYY88 pKa = 10.43QFLWCPTARR97 pKa = 11.84PGEE100 pKa = 4.27TSTGIDD106 pKa = 3.23GSAIDD111 pKa = 3.72KK112 pKa = 10.69AVRR115 pKa = 11.84TNTNCFMRR123 pKa = 11.84GVKK126 pKa = 10.23EE127 pKa = 4.04NVSLRR132 pKa = 11.84TNSGIAWRR140 pKa = 11.84WRR142 pKa = 11.84RR143 pKa = 11.84ICFTMKK149 pKa = 10.67GDD151 pKa = 3.76TLNFGDD157 pKa = 4.98NDD159 pKa = 4.41PDD161 pKa = 3.63TSNVARR167 pKa = 11.84QTSNGMMRR175 pKa = 11.84LVHH178 pKa = 6.46QDD180 pKa = 2.8LATIDD185 pKa = 4.19RR186 pKa = 11.84VTQVVFRR193 pKa = 11.84GQNQTDD199 pKa = 3.65WVNHH203 pKa = 6.7FIAPIDD209 pKa = 3.51TRR211 pKa = 11.84KK212 pKa = 9.85ISLRR216 pKa = 11.84FDD218 pKa = 3.28RR219 pKa = 11.84TRR221 pKa = 11.84VIQSNNDD228 pKa = 3.18SGTVRR233 pKa = 11.84YY234 pKa = 9.79YY235 pKa = 11.29KK236 pKa = 10.61DD237 pKa = 2.77WFAMNKK243 pKa = 9.54NIYY246 pKa = 10.45YY247 pKa = 10.21DD248 pKa = 3.7DD249 pKa = 4.22QEE251 pKa = 6.01NGDD254 pKa = 3.52QTFPSMFSVEE264 pKa = 4.28NKK266 pKa = 10.09QGMGDD271 pKa = 3.58YY272 pKa = 10.29YY273 pKa = 10.03IYY275 pKa = 10.43DD276 pKa = 3.79IIEE279 pKa = 4.3PQGGDD284 pKa = 3.2STDD287 pKa = 3.85AIEE290 pKa = 5.09FEE292 pKa = 4.43PQARR296 pKa = 11.84LYY298 pKa = 9.1WHH300 pKa = 7.04EE301 pKa = 4.13KK302 pKa = 9.24
MM1 pKa = 7.54AYY3 pKa = 10.05SRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84YY8 pKa = 8.43SAPRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84FRR16 pKa = 11.84RR17 pKa = 11.84GFKK20 pKa = 10.05RR21 pKa = 11.84RR22 pKa = 11.84SSRR25 pKa = 11.84LAYY28 pKa = 9.67RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84PYY34 pKa = 10.04VRR36 pKa = 11.84RR37 pKa = 11.84PYY39 pKa = 10.54RR40 pKa = 11.84KK41 pKa = 8.49RR42 pKa = 11.84TWTTRR47 pKa = 11.84AILNKK52 pKa = 9.05TSKK55 pKa = 10.61KK56 pKa = 9.44KK57 pKa = 9.17TDD59 pKa = 3.39TMQVWSDD66 pKa = 3.3TQTPGTYY73 pKa = 9.91VPGGAVIPAGTGAVYY88 pKa = 10.43QFLWCPTARR97 pKa = 11.84PGEE100 pKa = 4.27TSTGIDD106 pKa = 3.23GSAIDD111 pKa = 3.72KK112 pKa = 10.69AVRR115 pKa = 11.84TNTNCFMRR123 pKa = 11.84GVKK126 pKa = 10.23EE127 pKa = 4.04NVSLRR132 pKa = 11.84TNSGIAWRR140 pKa = 11.84WRR142 pKa = 11.84RR143 pKa = 11.84ICFTMKK149 pKa = 10.67GDD151 pKa = 3.76TLNFGDD157 pKa = 4.98NDD159 pKa = 4.41PDD161 pKa = 3.63TSNVARR167 pKa = 11.84QTSNGMMRR175 pKa = 11.84LVHH178 pKa = 6.46QDD180 pKa = 2.8LATIDD185 pKa = 4.19RR186 pKa = 11.84VTQVVFRR193 pKa = 11.84GQNQTDD199 pKa = 3.65WVNHH203 pKa = 6.7FIAPIDD209 pKa = 3.51TRR211 pKa = 11.84KK212 pKa = 9.85ISLRR216 pKa = 11.84FDD218 pKa = 3.28RR219 pKa = 11.84TRR221 pKa = 11.84VIQSNNDD228 pKa = 3.18SGTVRR233 pKa = 11.84YY234 pKa = 9.79YY235 pKa = 11.29KK236 pKa = 10.61DD237 pKa = 2.77WFAMNKK243 pKa = 9.54NIYY246 pKa = 10.45YY247 pKa = 10.21DD248 pKa = 3.7DD249 pKa = 4.22QEE251 pKa = 6.01NGDD254 pKa = 3.52QTFPSMFSVEE264 pKa = 4.28NKK266 pKa = 10.09QGMGDD271 pKa = 3.58YY272 pKa = 10.29YY273 pKa = 10.03IYY275 pKa = 10.43DD276 pKa = 3.79IIEE279 pKa = 4.3PQGGDD284 pKa = 3.2STDD287 pKa = 3.85AIEE290 pKa = 5.09FEE292 pKa = 4.43PQARR296 pKa = 11.84LYY298 pKa = 9.1WHH300 pKa = 7.04EE301 pKa = 4.13KK302 pKa = 9.24
Molecular weight: 35.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
623 |
302 |
321 |
311.5 |
35.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.618 ± 0.008 | 1.766 ± 0.536 |
7.384 ± 0.069 | 4.013 ± 0.947 |
5.618 ± 0.912 | 8.186 ± 0.856 |
1.926 ± 0.648 | 4.815 ± 0.335 |
3.852 ± 0.544 | 5.618 ± 1.832 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.247 ± 0.509 | 4.334 ± 0.899 |
4.655 ± 0.243 | 3.852 ± 0.544 |
9.631 ± 1.82 | 6.421 ± 0.32 |
6.742 ± 1.757 | 5.939 ± 0.215 |
2.889 ± 0.167 | 4.494 ± 0.328 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
