
Lactobacillus cacaonum DSM 21116
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Liquorilactobacillus; Liquorilactobacillus cacaonum
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
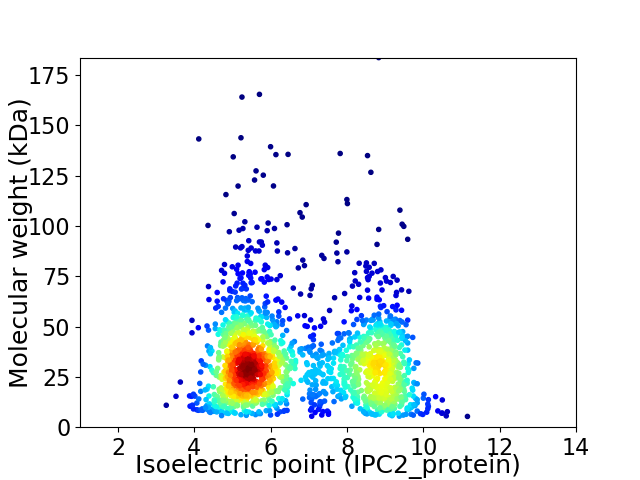
Virtual 2D-PAGE plot for 1801 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0R2CJT3|A0A0R2CJT3_9LACO Ion Mg(2+) C(O2+) transport protein OS=Lactobacillus cacaonum DSM 21116 OX=1423729 GN=FC80_GL000079 PE=4 SV=1
MM1 pKa = 7.37NLGSNPNGADD11 pKa = 3.65GIGFAFHH18 pKa = 7.1NGNTDD23 pKa = 3.97DD24 pKa = 4.75IGNSGANLGIGGLQNAIGFKK44 pKa = 10.62LDD46 pKa = 3.08TWKK49 pKa = 10.98NGYY52 pKa = 7.3QTPNKK57 pKa = 9.42DD58 pKa = 3.19VPGAQISTPDD68 pKa = 3.37SNGFGWNGDD77 pKa = 3.82SKK79 pKa = 9.98GTPYY83 pKa = 10.1GTFVTTSDD91 pKa = 3.62QQIKK95 pKa = 7.23TTDD98 pKa = 3.2GTEE101 pKa = 3.99VQRR104 pKa = 11.84WWAVDD109 pKa = 3.02SGTAQSLSKK118 pKa = 10.99ADD120 pKa = 3.22IDD122 pKa = 4.63GNFHH126 pKa = 6.35NFVVSYY132 pKa = 10.69DD133 pKa = 3.2GSTRR137 pKa = 11.84TLTINYY143 pKa = 7.85TEE145 pKa = 4.36TDD147 pKa = 3.21GTMLTWSMQVDD158 pKa = 3.85DD159 pKa = 5.78AYY161 pKa = 9.93TAMSMIVSASTGSATNLQQFKK182 pKa = 10.26IDD184 pKa = 3.51SFDD187 pKa = 3.55FQEE190 pKa = 4.19AATVNVKK197 pKa = 10.5YY198 pKa = 10.73VDD200 pKa = 3.62EE201 pKa = 4.89NGNSIADD208 pKa = 3.95GEE210 pKa = 4.62VTYY213 pKa = 10.82PDD215 pKa = 3.55GPYY218 pKa = 11.03VNGTYY223 pKa = 7.49TTVQKK228 pKa = 10.57DD229 pKa = 3.09IPNYY233 pKa = 10.18KK234 pKa = 10.01FIGMGDD240 pKa = 3.36NATTGVSIAKK250 pKa = 9.8DD251 pKa = 3.17GTMADD256 pKa = 4.08DD257 pKa = 4.56GNNGTVIYY265 pKa = 9.71VYY267 pKa = 10.88APAYY271 pKa = 10.06SATSKK276 pKa = 9.9TINEE280 pKa = 4.46TINYY284 pKa = 8.98VDD286 pKa = 3.37QNGNKK291 pKa = 9.47VADD294 pKa = 4.45SYY296 pKa = 10.95TATPITVVTVTNPVDD311 pKa = 3.59GSKK314 pKa = 9.54TYY316 pKa = 10.31YY317 pKa = 10.02YY318 pKa = 11.02KK319 pKa = 10.78SGTDD323 pKa = 3.46DD324 pKa = 4.68TPTLDD329 pKa = 3.51NNGVPEE335 pKa = 4.39GEE337 pKa = 4.37GWTKK341 pKa = 11.14ADD343 pKa = 3.65SSDD346 pKa = 3.18FTAVKK351 pKa = 10.55NPTVDD356 pKa = 3.42GYY358 pKa = 11.73KK359 pKa = 10.35VISNDD364 pKa = 3.78APNSDD369 pKa = 3.82LTQVADD375 pKa = 3.57QTITADD381 pKa = 4.32SSDD384 pKa = 3.47LNYY387 pKa = 10.2TVVYY391 pKa = 10.14APAYY395 pKa = 9.39SATSKK400 pKa = 9.89TINEE404 pKa = 4.12TXXXXNTSNTSNTNTSNTNTNTTDD428 pKa = 3.02ANEE431 pKa = 4.06YY432 pKa = 10.86EE433 pKa = 4.3RR434 pKa = 11.84FKK436 pKa = 11.33CGNTKK441 pKa = 10.5GNEE444 pKa = 3.79
MM1 pKa = 7.37NLGSNPNGADD11 pKa = 3.65GIGFAFHH18 pKa = 7.1NGNTDD23 pKa = 3.97DD24 pKa = 4.75IGNSGANLGIGGLQNAIGFKK44 pKa = 10.62LDD46 pKa = 3.08TWKK49 pKa = 10.98NGYY52 pKa = 7.3QTPNKK57 pKa = 9.42DD58 pKa = 3.19VPGAQISTPDD68 pKa = 3.37SNGFGWNGDD77 pKa = 3.82SKK79 pKa = 9.98GTPYY83 pKa = 10.1GTFVTTSDD91 pKa = 3.62QQIKK95 pKa = 7.23TTDD98 pKa = 3.2GTEE101 pKa = 3.99VQRR104 pKa = 11.84WWAVDD109 pKa = 3.02SGTAQSLSKK118 pKa = 10.99ADD120 pKa = 3.22IDD122 pKa = 4.63GNFHH126 pKa = 6.35NFVVSYY132 pKa = 10.69DD133 pKa = 3.2GSTRR137 pKa = 11.84TLTINYY143 pKa = 7.85TEE145 pKa = 4.36TDD147 pKa = 3.21GTMLTWSMQVDD158 pKa = 3.85DD159 pKa = 5.78AYY161 pKa = 9.93TAMSMIVSASTGSATNLQQFKK182 pKa = 10.26IDD184 pKa = 3.51SFDD187 pKa = 3.55FQEE190 pKa = 4.19AATVNVKK197 pKa = 10.5YY198 pKa = 10.73VDD200 pKa = 3.62EE201 pKa = 4.89NGNSIADD208 pKa = 3.95GEE210 pKa = 4.62VTYY213 pKa = 10.82PDD215 pKa = 3.55GPYY218 pKa = 11.03VNGTYY223 pKa = 7.49TTVQKK228 pKa = 10.57DD229 pKa = 3.09IPNYY233 pKa = 10.18KK234 pKa = 10.01FIGMGDD240 pKa = 3.36NATTGVSIAKK250 pKa = 9.8DD251 pKa = 3.17GTMADD256 pKa = 4.08DD257 pKa = 4.56GNNGTVIYY265 pKa = 9.71VYY267 pKa = 10.88APAYY271 pKa = 10.06SATSKK276 pKa = 9.9TINEE280 pKa = 4.46TINYY284 pKa = 8.98VDD286 pKa = 3.37QNGNKK291 pKa = 9.47VADD294 pKa = 4.45SYY296 pKa = 10.95TATPITVVTVTNPVDD311 pKa = 3.59GSKK314 pKa = 9.54TYY316 pKa = 10.31YY317 pKa = 10.02YY318 pKa = 11.02KK319 pKa = 10.78SGTDD323 pKa = 3.46DD324 pKa = 4.68TPTLDD329 pKa = 3.51NNGVPEE335 pKa = 4.39GEE337 pKa = 4.37GWTKK341 pKa = 11.14ADD343 pKa = 3.65SSDD346 pKa = 3.18FTAVKK351 pKa = 10.55NPTVDD356 pKa = 3.42GYY358 pKa = 11.73KK359 pKa = 10.35VISNDD364 pKa = 3.78APNSDD369 pKa = 3.82LTQVADD375 pKa = 3.57QTITADD381 pKa = 4.32SSDD384 pKa = 3.47LNYY387 pKa = 10.2TVVYY391 pKa = 10.14APAYY395 pKa = 9.39SATSKK400 pKa = 9.89TINEE404 pKa = 4.12TXXXXNTSNTSNTNTSNTNTNTTDD428 pKa = 3.02ANEE431 pKa = 4.06YY432 pKa = 10.86EE433 pKa = 4.3RR434 pKa = 11.84FKK436 pKa = 11.33CGNTKK441 pKa = 10.5GNEE444 pKa = 3.79
Molecular weight: 46.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0R2CDN9|A0A0R2CDN9_9LACO ABC transporter ATP-binding protein OS=Lactobacillus cacaonum DSM 21116 OX=1423729 GN=FC80_GL001724 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.69KK9 pKa = 7.77RR10 pKa = 11.84HH11 pKa = 4.99RR12 pKa = 11.84QRR14 pKa = 11.84VHH16 pKa = 6.07GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.69QPKK8 pKa = 8.69KK9 pKa = 7.77RR10 pKa = 11.84HH11 pKa = 4.99RR12 pKa = 11.84QRR14 pKa = 11.84VHH16 pKa = 6.07GFRR19 pKa = 11.84KK20 pKa = 10.03RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.05GRR39 pKa = 11.84KK40 pKa = 8.7VLSAA44 pKa = 4.05
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
557821 |
44 |
1707 |
309.7 |
34.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.908 ± 0.053 | 0.554 ± 0.013 |
5.259 ± 0.059 | 6.392 ± 0.075 |
4.648 ± 0.057 | 6.493 ± 0.052 |
1.74 ± 0.022 | 8.34 ± 0.064 |
7.762 ± 0.054 | 9.878 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.553 ± 0.027 | 5.43 ± 0.043 |
3.108 ± 0.034 | 3.814 ± 0.037 |
3.64 ± 0.038 | 6.481 ± 0.059 |
5.657 ± 0.048 | 6.903 ± 0.045 |
0.876 ± 0.019 | 3.561 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
