
Hymenobacter gelipurpurascens
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Hymenobacteraceae; Hymenobacter
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
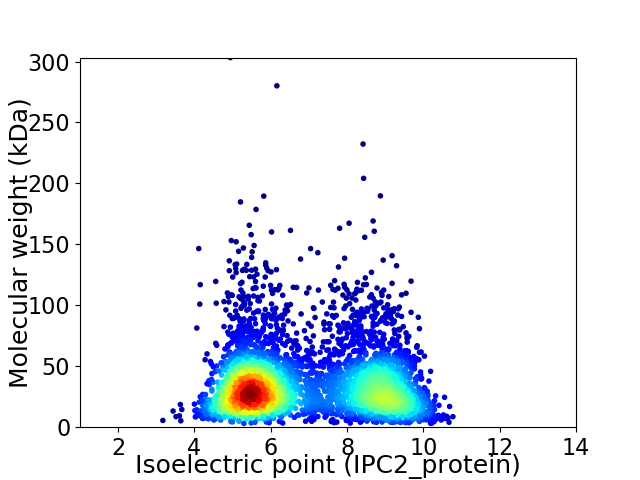
Virtual 2D-PAGE plot for 4241 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
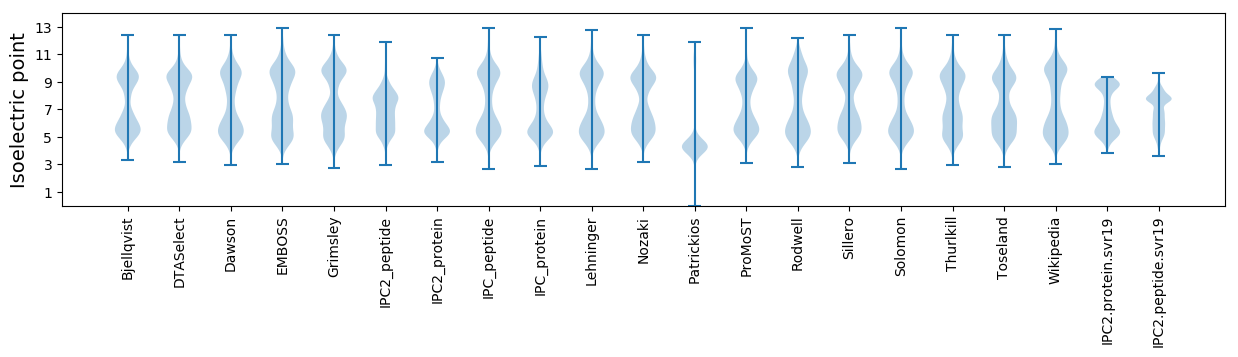
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A212T2X0|A0A212T2X0_9BACT Short-chain dehydrogenase OS=Hymenobacter gelipurpurascens OX=89968 GN=SAMN06265337_0241 PE=3 SV=1
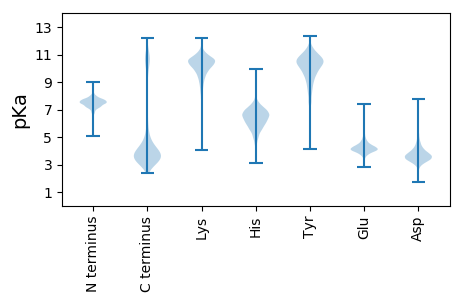
MM1 pKa = 8.43DD2 pKa = 3.81YY3 pKa = 11.36TNDD6 pKa = 3.5PEE8 pKa = 5.64LNGKK12 pKa = 8.58YY13 pKa = 10.13LGTITKK19 pKa = 10.4DD20 pKa = 3.03FATVSDD26 pKa = 4.14TLSEE30 pKa = 4.07ASSQIRR36 pKa = 11.84KK37 pKa = 9.39RR38 pKa = 11.84DD39 pKa = 3.18ISKK42 pKa = 10.51YY43 pKa = 9.21PIFVFARR50 pKa = 11.84QEE52 pKa = 4.02VPLGGLLINADD63 pKa = 3.75EE64 pKa = 5.58ANLEE68 pKa = 4.0WHH70 pKa = 6.32VFASYY75 pKa = 11.36LEE77 pKa = 4.91LFVQQGIVSQEE88 pKa = 4.01GIEE91 pKa = 4.42AFQSTYY97 pKa = 10.62RR98 pKa = 11.84DD99 pKa = 3.27ADD101 pKa = 3.84EE102 pKa = 4.42YY103 pKa = 11.61CCLFVLDD110 pKa = 4.67EE111 pKa = 4.36EE112 pKa = 4.73FTNFVYY118 pKa = 10.29IPYY121 pKa = 10.15PEE123 pKa = 4.46DD124 pKa = 3.11
MM1 pKa = 8.43DD2 pKa = 3.81YY3 pKa = 11.36TNDD6 pKa = 3.5PEE8 pKa = 5.64LNGKK12 pKa = 8.58YY13 pKa = 10.13LGTITKK19 pKa = 10.4DD20 pKa = 3.03FATVSDD26 pKa = 4.14TLSEE30 pKa = 4.07ASSQIRR36 pKa = 11.84KK37 pKa = 9.39RR38 pKa = 11.84DD39 pKa = 3.18ISKK42 pKa = 10.51YY43 pKa = 9.21PIFVFARR50 pKa = 11.84QEE52 pKa = 4.02VPLGGLLINADD63 pKa = 3.75EE64 pKa = 5.58ANLEE68 pKa = 4.0WHH70 pKa = 6.32VFASYY75 pKa = 11.36LEE77 pKa = 4.91LFVQQGIVSQEE88 pKa = 4.01GIEE91 pKa = 4.42AFQSTYY97 pKa = 10.62RR98 pKa = 11.84DD99 pKa = 3.27ADD101 pKa = 3.84EE102 pKa = 4.42YY103 pKa = 11.61CCLFVLDD110 pKa = 4.67EE111 pKa = 4.36EE112 pKa = 4.73FTNFVYY118 pKa = 10.29IPYY121 pKa = 10.15PEE123 pKa = 4.46DD124 pKa = 3.11
Molecular weight: 14.23 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A212UH86|A0A212UH86_9BACT Periplasmic chaperone for outer membrane proteins Skp OS=Hymenobacter gelipurpurascens OX=89968 GN=SAMN06265337_4018 PE=3 SV=1
MM1 pKa = 7.34AHH3 pKa = 7.5DD4 pKa = 3.84GTLGHH9 pKa = 6.94FLPFLLGRR17 pKa = 11.84LLRR20 pKa = 11.84AVLWPDD26 pKa = 3.08AGEE29 pKa = 4.19GRR31 pKa = 11.84AALIFLTLFCILYY44 pKa = 10.0GVVLAFMLNNEE55 pKa = 4.2NGIPSGFAQKK65 pKa = 11.02LLFALNGILYY75 pKa = 10.47ASALLVDD82 pKa = 5.36FVPTYY87 pKa = 10.84RR88 pKa = 11.84PVQPPLPDD96 pKa = 3.65HH97 pKa = 6.6FPVSGRR103 pKa = 11.84LNLITAFLLDD113 pKa = 5.42LITVRR118 pKa = 11.84RR119 pKa = 11.84FLLLLFLLVALACAPHH135 pKa = 6.13SWRR138 pKa = 11.84PLGLNLLVLLSAGAASFNLRR158 pKa = 11.84LLLSMGRR165 pKa = 11.84WRR167 pKa = 11.84HH168 pKa = 5.91LLFALNVLCLAAAAGWLSTFISLPLAATALVVVAIAGPLVLWGVALAGLGEE219 pKa = 4.43KK220 pKa = 10.48FNARR224 pKa = 11.84HH225 pKa = 5.79LAVAPEE231 pKa = 4.4SKK233 pKa = 10.57SEE235 pKa = 3.93NQLLARR241 pKa = 11.84LSPEE245 pKa = 2.99WKK247 pKa = 10.2AYY249 pKa = 9.69LRR251 pKa = 11.84KK252 pKa = 9.59CWPALSVALVAKK264 pKa = 10.03LVILGLSSQMTATRR278 pKa = 11.84HH279 pKa = 5.36GDD281 pKa = 3.41PLNGVFWMSFLPIISFTYY299 pKa = 10.35VNNNLFGIIGPVSANEE315 pKa = 3.87LARR318 pKa = 11.84LGLTKK323 pKa = 10.49RR324 pKa = 11.84LLYY327 pKa = 10.59LYY329 pKa = 9.9LRR331 pKa = 11.84LVGPVLLLDD340 pKa = 4.2CLLSVVVLLALFPQQRR356 pKa = 11.84WSLIGLIPLAAVAMLALGLWGSLYY380 pKa = 10.08KK381 pKa = 10.8AKK383 pKa = 10.5AISKK387 pKa = 10.55SIDD390 pKa = 3.42LANMRR395 pKa = 11.84NNASTLMNLLSIGVGAALFFMPWWWARR422 pKa = 11.84IVLAIVVMASAWWPVQRR439 pKa = 11.84VLRR442 pKa = 11.84NEE444 pKa = 3.95GGLRR448 pKa = 11.84RR449 pKa = 11.84QLWGALNSS457 pKa = 3.82
MM1 pKa = 7.34AHH3 pKa = 7.5DD4 pKa = 3.84GTLGHH9 pKa = 6.94FLPFLLGRR17 pKa = 11.84LLRR20 pKa = 11.84AVLWPDD26 pKa = 3.08AGEE29 pKa = 4.19GRR31 pKa = 11.84AALIFLTLFCILYY44 pKa = 10.0GVVLAFMLNNEE55 pKa = 4.2NGIPSGFAQKK65 pKa = 11.02LLFALNGILYY75 pKa = 10.47ASALLVDD82 pKa = 5.36FVPTYY87 pKa = 10.84RR88 pKa = 11.84PVQPPLPDD96 pKa = 3.65HH97 pKa = 6.6FPVSGRR103 pKa = 11.84LNLITAFLLDD113 pKa = 5.42LITVRR118 pKa = 11.84RR119 pKa = 11.84FLLLLFLLVALACAPHH135 pKa = 6.13SWRR138 pKa = 11.84PLGLNLLVLLSAGAASFNLRR158 pKa = 11.84LLLSMGRR165 pKa = 11.84WRR167 pKa = 11.84HH168 pKa = 5.91LLFALNVLCLAAAAGWLSTFISLPLAATALVVVAIAGPLVLWGVALAGLGEE219 pKa = 4.43KK220 pKa = 10.48FNARR224 pKa = 11.84HH225 pKa = 5.79LAVAPEE231 pKa = 4.4SKK233 pKa = 10.57SEE235 pKa = 3.93NQLLARR241 pKa = 11.84LSPEE245 pKa = 2.99WKK247 pKa = 10.2AYY249 pKa = 9.69LRR251 pKa = 11.84KK252 pKa = 9.59CWPALSVALVAKK264 pKa = 10.03LVILGLSSQMTATRR278 pKa = 11.84HH279 pKa = 5.36GDD281 pKa = 3.41PLNGVFWMSFLPIISFTYY299 pKa = 10.35VNNNLFGIIGPVSANEE315 pKa = 3.87LARR318 pKa = 11.84LGLTKK323 pKa = 10.49RR324 pKa = 11.84LLYY327 pKa = 10.59LYY329 pKa = 9.9LRR331 pKa = 11.84LVGPVLLLDD340 pKa = 4.2CLLSVVVLLALFPQQRR356 pKa = 11.84WSLIGLIPLAAVAMLALGLWGSLYY380 pKa = 10.08KK381 pKa = 10.8AKK383 pKa = 10.5AISKK387 pKa = 10.55SIDD390 pKa = 3.42LANMRR395 pKa = 11.84NNASTLMNLLSIGVGAALFFMPWWWARR422 pKa = 11.84IVLAIVVMASAWWPVQRR439 pKa = 11.84VLRR442 pKa = 11.84NEE444 pKa = 3.95GGLRR448 pKa = 11.84RR449 pKa = 11.84QLWGALNSS457 pKa = 3.82
Molecular weight: 49.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1451777 |
29 |
3034 |
342.3 |
37.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.929 ± 0.046 | 0.708 ± 0.011 |
4.835 ± 0.026 | 5.493 ± 0.041 |
4.13 ± 0.024 | 7.507 ± 0.034 |
2.106 ± 0.019 | 4.607 ± 0.031 |
4.236 ± 0.037 | 10.992 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.0 ± 0.017 | 3.793 ± 0.035 |
5.106 ± 0.026 | 4.848 ± 0.032 |
5.873 ± 0.03 | 5.727 ± 0.032 |
6.236 ± 0.041 | 7.096 ± 0.028 |
1.246 ± 0.017 | 3.533 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
