
Ruminiclostridium sufflavum DSM 19573
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Ruminiclostridium; Ruminiclostridium sufflavum
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
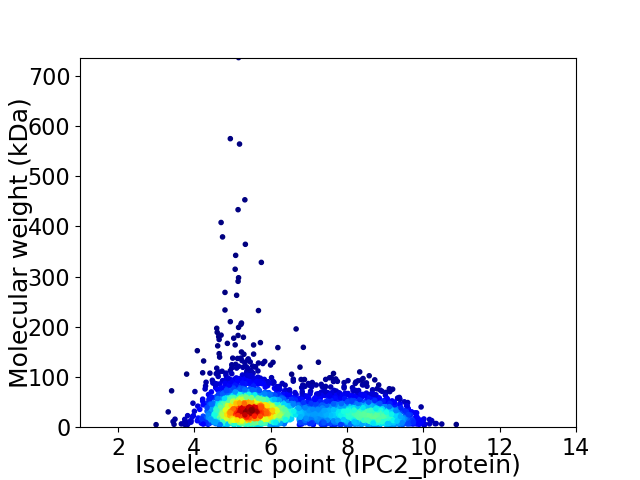
Virtual 2D-PAGE plot for 3674 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A318XSA3|A0A318XSA3_9FIRM N-acylneuraminate cytidylyltransferase OS=Ruminiclostridium sufflavum DSM 19573 OX=1121337 GN=LY28_00944 PE=4 SV=1
MM1 pKa = 7.47LKK3 pKa = 10.03KK4 pKa = 10.4SKK6 pKa = 10.64SILFIILITIALIPSITFVQAVEE29 pKa = 4.79PIQGGTSINNAALISKK45 pKa = 8.38TDD47 pKa = 3.54SLISITGDD55 pKa = 3.12SINPVNAYY63 pKa = 9.86FKK65 pKa = 10.64FVAATTGNFTITTSPVSADD84 pKa = 3.63TEE86 pKa = 4.05AMIYY90 pKa = 10.47YY91 pKa = 7.77DD92 pKa = 3.47TSEE95 pKa = 4.22TSGLQAIGDD104 pKa = 3.83NTDD107 pKa = 4.25CEE109 pKa = 5.07FLLQDD114 pKa = 4.6NLNEE118 pKa = 3.99GQIYY122 pKa = 8.69YY123 pKa = 10.63LKK125 pKa = 10.03IWEE128 pKa = 4.35YY129 pKa = 11.29ANGNAEE135 pKa = 3.91VTINITGGLLMAEE148 pKa = 4.27PEE150 pKa = 3.99ITITNSAANLSYY162 pKa = 10.38MGGQKK167 pKa = 9.79ISISGTVFDD176 pKa = 4.88AEE178 pKa = 4.91GYY180 pKa = 10.25DD181 pKa = 3.53VTVSATIDD189 pKa = 3.2GHH191 pKa = 6.52LVSKK195 pKa = 9.02TVSGGAGAWTLEE207 pKa = 3.59WDD209 pKa = 3.66IDD211 pKa = 4.43SMNISDD217 pKa = 4.87NKK219 pKa = 8.07YY220 pKa = 9.74QNIEE224 pKa = 4.47FIVDD228 pKa = 3.37DD229 pKa = 4.4GTPVGPVSAIHH240 pKa = 6.28TGDD243 pKa = 3.06IVVDD247 pKa = 3.81KK248 pKa = 9.59TPPPAPTAVTVTPVGGMVIANSLNKK273 pKa = 10.47SNTNMTATATITAAEE288 pKa = 4.04AAGGKK293 pKa = 10.13AEE295 pKa = 4.68LYY297 pKa = 10.79LDD299 pKa = 4.1GVLLATDD306 pKa = 3.59NSIASEE312 pKa = 4.06DD313 pKa = 3.88TQVTFDD319 pKa = 4.54LGKK322 pKa = 9.01STNAEE327 pKa = 3.63LQAAVVSSGTVSVKK341 pKa = 10.74LYY343 pKa = 10.54DD344 pKa = 3.52AAGNSSTSSVNNPVLAVDD362 pKa = 3.9YY363 pKa = 10.49NISEE367 pKa = 4.39QVPPTATVYY376 pKa = 10.58TIGGTLRR383 pKa = 11.84VGGTLTGSYY392 pKa = 9.53TYY394 pKa = 11.22YY395 pKa = 10.75DD396 pKa = 3.59ANGDD400 pKa = 3.82SEE402 pKa = 5.08GISTFKK408 pKa = 10.33WYY410 pKa = 8.52TADD413 pKa = 5.45DD414 pKa = 3.74SSGTNKK420 pKa = 9.78TEE422 pKa = 3.73IAGANAEE429 pKa = 4.46SYY431 pKa = 11.15VLTSADD437 pKa = 2.83IGKK440 pKa = 10.27YY441 pKa = 9.68ISFEE445 pKa = 4.29VKK447 pKa = 9.93PVALTGTTQGTAA459 pKa = 3.01
MM1 pKa = 7.47LKK3 pKa = 10.03KK4 pKa = 10.4SKK6 pKa = 10.64SILFIILITIALIPSITFVQAVEE29 pKa = 4.79PIQGGTSINNAALISKK45 pKa = 8.38TDD47 pKa = 3.54SLISITGDD55 pKa = 3.12SINPVNAYY63 pKa = 9.86FKK65 pKa = 10.64FVAATTGNFTITTSPVSADD84 pKa = 3.63TEE86 pKa = 4.05AMIYY90 pKa = 10.47YY91 pKa = 7.77DD92 pKa = 3.47TSEE95 pKa = 4.22TSGLQAIGDD104 pKa = 3.83NTDD107 pKa = 4.25CEE109 pKa = 5.07FLLQDD114 pKa = 4.6NLNEE118 pKa = 3.99GQIYY122 pKa = 8.69YY123 pKa = 10.63LKK125 pKa = 10.03IWEE128 pKa = 4.35YY129 pKa = 11.29ANGNAEE135 pKa = 3.91VTINITGGLLMAEE148 pKa = 4.27PEE150 pKa = 3.99ITITNSAANLSYY162 pKa = 10.38MGGQKK167 pKa = 9.79ISISGTVFDD176 pKa = 4.88AEE178 pKa = 4.91GYY180 pKa = 10.25DD181 pKa = 3.53VTVSATIDD189 pKa = 3.2GHH191 pKa = 6.52LVSKK195 pKa = 9.02TVSGGAGAWTLEE207 pKa = 3.59WDD209 pKa = 3.66IDD211 pKa = 4.43SMNISDD217 pKa = 4.87NKK219 pKa = 8.07YY220 pKa = 9.74QNIEE224 pKa = 4.47FIVDD228 pKa = 3.37DD229 pKa = 4.4GTPVGPVSAIHH240 pKa = 6.28TGDD243 pKa = 3.06IVVDD247 pKa = 3.81KK248 pKa = 9.59TPPPAPTAVTVTPVGGMVIANSLNKK273 pKa = 10.47SNTNMTATATITAAEE288 pKa = 4.04AAGGKK293 pKa = 10.13AEE295 pKa = 4.68LYY297 pKa = 10.79LDD299 pKa = 4.1GVLLATDD306 pKa = 3.59NSIASEE312 pKa = 4.06DD313 pKa = 3.88TQVTFDD319 pKa = 4.54LGKK322 pKa = 9.01STNAEE327 pKa = 3.63LQAAVVSSGTVSVKK341 pKa = 10.74LYY343 pKa = 10.54DD344 pKa = 3.52AAGNSSTSSVNNPVLAVDD362 pKa = 3.9YY363 pKa = 10.49NISEE367 pKa = 4.39QVPPTATVYY376 pKa = 10.58TIGGTLRR383 pKa = 11.84VGGTLTGSYY392 pKa = 9.53TYY394 pKa = 11.22YY395 pKa = 10.75DD396 pKa = 3.59ANGDD400 pKa = 3.82SEE402 pKa = 5.08GISTFKK408 pKa = 10.33WYY410 pKa = 8.52TADD413 pKa = 5.45DD414 pKa = 3.74SSGTNKK420 pKa = 9.78TEE422 pKa = 3.73IAGANAEE429 pKa = 4.46SYY431 pKa = 11.15VLTSADD437 pKa = 2.83IGKK440 pKa = 10.27YY441 pKa = 9.68ISFEE445 pKa = 4.29VKK447 pKa = 9.93PVALTGTTQGTAA459 pKa = 3.01
Molecular weight: 47.81 kDa
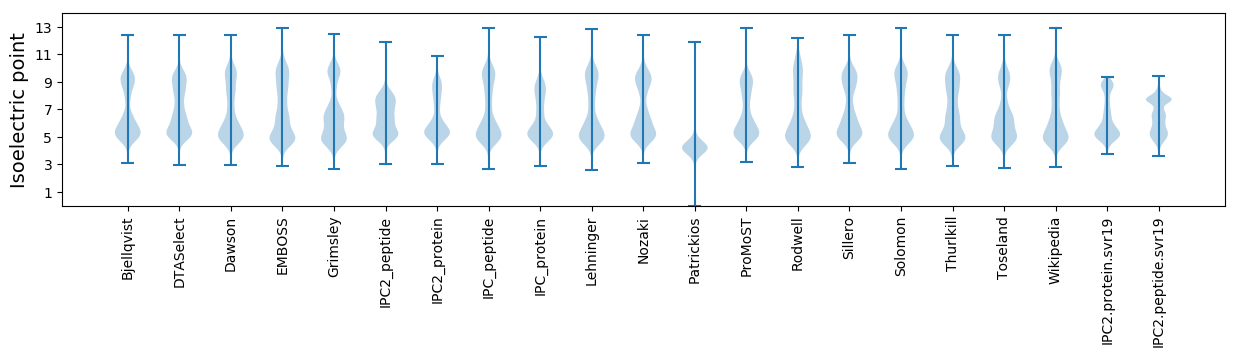
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A318Y9S7|A0A318Y9S7_9FIRM ATP-binding cassette subfamily F protein uup OS=Ruminiclostridium sufflavum DSM 19573 OX=1121337 GN=LY28_00966 PE=4 SV=1
MM1 pKa = 7.45LRR3 pKa = 11.84TYY5 pKa = 9.82QPKK8 pKa = 9.61KK9 pKa = 8.53RR10 pKa = 11.84HH11 pKa = 5.03RR12 pKa = 11.84QKK14 pKa = 10.33EE15 pKa = 3.73HH16 pKa = 5.99GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 7.2VLRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.24GRR39 pKa = 11.84KK40 pKa = 9.08AISAA44 pKa = 3.87
MM1 pKa = 7.45LRR3 pKa = 11.84TYY5 pKa = 9.82QPKK8 pKa = 9.61KK9 pKa = 8.53RR10 pKa = 11.84HH11 pKa = 5.03RR12 pKa = 11.84QKK14 pKa = 10.33EE15 pKa = 3.73HH16 pKa = 5.99GFRR19 pKa = 11.84KK20 pKa = 10.01RR21 pKa = 11.84MSTSNGRR28 pKa = 11.84KK29 pKa = 7.2VLRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.24GRR39 pKa = 11.84KK40 pKa = 9.08AISAA44 pKa = 3.87
Molecular weight: 5.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1258753 |
39 |
6533 |
342.6 |
38.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.166 ± 0.047 | 1.36 ± 0.018 |
5.518 ± 0.034 | 6.924 ± 0.043 |
4.252 ± 0.035 | 6.845 ± 0.043 |
1.401 ± 0.017 | 8.691 ± 0.049 |
7.627 ± 0.042 | 8.836 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.632 ± 0.021 | 5.355 ± 0.034 |
3.153 ± 0.046 | 2.85 ± 0.022 |
3.886 ± 0.03 | 6.788 ± 0.033 |
5.223 ± 0.048 | 6.495 ± 0.039 |
0.862 ± 0.016 | 4.137 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
