
Aliifodinibius halophilus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Balneolaeota; Balneolia; Balneolales; Balneolaceae; Aliifodinibius
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
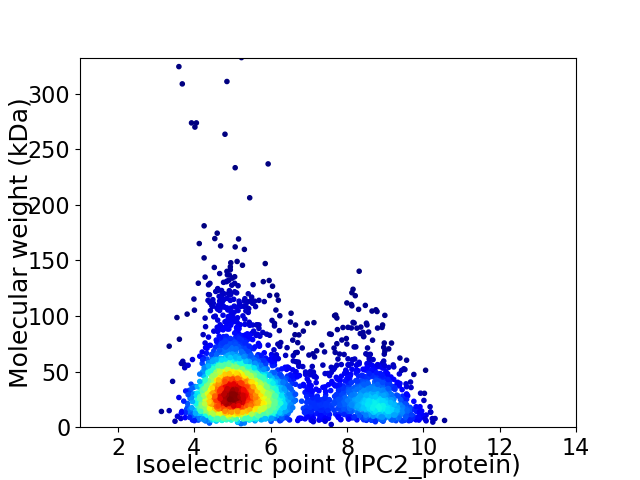
Virtual 2D-PAGE plot for 3590 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M1T1B1|A0A6M1T1B1_9BACT Indole-3-glycerol phosphate synthase OS=Aliifodinibius halophilus OX=1736908 GN=trpC PE=3 SV=1
MM1 pKa = 7.47KK2 pKa = 10.65LSFTKK7 pKa = 10.48VASALVLALTVIVTGCNDD25 pKa = 4.41DD26 pKa = 4.31GPNGPDD32 pKa = 3.41SNPLSVEE39 pKa = 4.41SINPTAGQPGDD50 pKa = 3.61EE51 pKa = 4.25VTITGTGFANNADD64 pKa = 4.1DD65 pKa = 5.01NSVTFAGDD73 pKa = 3.14VTASISSVSATEE85 pKa = 3.99LVVTVPQGATTGPITVSVDD104 pKa = 3.16GEE106 pKa = 4.41SATTSEE112 pKa = 4.63FAISSEE118 pKa = 4.03PQPEE122 pKa = 4.01IPEE125 pKa = 4.63PYY127 pKa = 10.5ASFPGTKK134 pKa = 9.11IAADD138 pKa = 3.96EE139 pKa = 4.28TWSNDD144 pKa = 2.94TTLVGPHH151 pKa = 5.95FVLPGVTLTVEE162 pKa = 4.2AGVEE166 pKa = 4.23VTFEE170 pKa = 3.98YY171 pKa = 11.07HH172 pKa = 6.48NGNDD176 pKa = 3.81DD177 pKa = 3.69KK178 pKa = 11.79VGAIYY183 pKa = 9.84TLASDD188 pKa = 4.26DD189 pKa = 4.02QNFSEE194 pKa = 4.41PRR196 pKa = 11.84EE197 pKa = 4.14TAKK200 pKa = 10.84LVAEE204 pKa = 4.95GTADD208 pKa = 3.42QPIVFTSDD216 pKa = 2.62RR217 pKa = 11.84KK218 pKa = 10.5QINSWGGIIMTGRR231 pKa = 11.84ASNNIPGGQGTVEE244 pKa = 5.02GIEE247 pKa = 3.63QDD249 pKa = 3.02IFYY252 pKa = 10.86GADD255 pKa = 3.18ADD257 pKa = 5.1GNFNDD262 pKa = 4.63SDD264 pKa = 4.18DD265 pKa = 4.34SGSLEE270 pKa = 4.03YY271 pKa = 11.15VRR273 pKa = 11.84IEE275 pKa = 3.8YY276 pKa = 10.48AGYY279 pKa = 9.92SINAGSEE286 pKa = 4.21LQSLTLYY293 pKa = 10.92SIGSEE298 pKa = 4.09TNINHH303 pKa = 5.52VNLFGSSDD311 pKa = 3.57DD312 pKa = 4.36GIEE315 pKa = 4.15LFGGTVDD322 pKa = 3.05IKK324 pKa = 11.11YY325 pKa = 10.12LVVYY329 pKa = 10.02GADD332 pKa = 4.11DD333 pKa = 4.4DD334 pKa = 5.52SFDD337 pKa = 4.69YY338 pKa = 11.19DD339 pKa = 3.78QGWSGKK345 pKa = 7.89GQFWLAVQRR354 pKa = 11.84DD355 pKa = 3.84GADD358 pKa = 2.9QGFEE362 pKa = 3.97NDD364 pKa = 3.56GCADD368 pKa = 4.62EE369 pKa = 5.55ADD371 pKa = 4.47CDD373 pKa = 3.82QGSGPGSPNISNATVIAPKK392 pKa = 9.15NTASGKK398 pKa = 10.64DD399 pKa = 3.45NMGLKK404 pKa = 10.17LRR406 pKa = 11.84EE407 pKa = 4.27GLQGAYY413 pKa = 10.46RR414 pKa = 11.84NIIVSNFVGNPFYY427 pKa = 11.2LDD429 pKa = 3.61GEE431 pKa = 4.48NNANDD436 pKa = 3.97DD437 pKa = 3.7TYY439 pKa = 11.79EE440 pKa = 4.42NYY442 pKa = 10.64DD443 pKa = 3.34SGDD446 pKa = 3.41LVLEE450 pKa = 4.15NFVLQNNGPWASSDD464 pKa = 3.16EE465 pKa = 4.23SRR467 pKa = 11.84YY468 pKa = 9.47SAVYY472 pKa = 9.98EE473 pKa = 4.11YY474 pKa = 10.76TSSDD478 pKa = 3.24KK479 pKa = 11.09PADD482 pKa = 3.71LFNNPSNYY490 pKa = 10.11DD491 pKa = 3.24YY492 pKa = 11.34SLPSGSAYY500 pKa = 10.66LSGGQAPSDD509 pKa = 3.31SWFDD513 pKa = 3.54DD514 pKa = 3.39VSFRR518 pKa = 11.84GAVGEE523 pKa = 6.14SGTQWDD529 pKa = 4.3WVNEE533 pKa = 4.69GIWVKK538 pKa = 10.39WPII541 pKa = 3.56
MM1 pKa = 7.47KK2 pKa = 10.65LSFTKK7 pKa = 10.48VASALVLALTVIVTGCNDD25 pKa = 4.41DD26 pKa = 4.31GPNGPDD32 pKa = 3.41SNPLSVEE39 pKa = 4.41SINPTAGQPGDD50 pKa = 3.61EE51 pKa = 4.25VTITGTGFANNADD64 pKa = 4.1DD65 pKa = 5.01NSVTFAGDD73 pKa = 3.14VTASISSVSATEE85 pKa = 3.99LVVTVPQGATTGPITVSVDD104 pKa = 3.16GEE106 pKa = 4.41SATTSEE112 pKa = 4.63FAISSEE118 pKa = 4.03PQPEE122 pKa = 4.01IPEE125 pKa = 4.63PYY127 pKa = 10.5ASFPGTKK134 pKa = 9.11IAADD138 pKa = 3.96EE139 pKa = 4.28TWSNDD144 pKa = 2.94TTLVGPHH151 pKa = 5.95FVLPGVTLTVEE162 pKa = 4.2AGVEE166 pKa = 4.23VTFEE170 pKa = 3.98YY171 pKa = 11.07HH172 pKa = 6.48NGNDD176 pKa = 3.81DD177 pKa = 3.69KK178 pKa = 11.79VGAIYY183 pKa = 9.84TLASDD188 pKa = 4.26DD189 pKa = 4.02QNFSEE194 pKa = 4.41PRR196 pKa = 11.84EE197 pKa = 4.14TAKK200 pKa = 10.84LVAEE204 pKa = 4.95GTADD208 pKa = 3.42QPIVFTSDD216 pKa = 2.62RR217 pKa = 11.84KK218 pKa = 10.5QINSWGGIIMTGRR231 pKa = 11.84ASNNIPGGQGTVEE244 pKa = 5.02GIEE247 pKa = 3.63QDD249 pKa = 3.02IFYY252 pKa = 10.86GADD255 pKa = 3.18ADD257 pKa = 5.1GNFNDD262 pKa = 4.63SDD264 pKa = 4.18DD265 pKa = 4.34SGSLEE270 pKa = 4.03YY271 pKa = 11.15VRR273 pKa = 11.84IEE275 pKa = 3.8YY276 pKa = 10.48AGYY279 pKa = 9.92SINAGSEE286 pKa = 4.21LQSLTLYY293 pKa = 10.92SIGSEE298 pKa = 4.09TNINHH303 pKa = 5.52VNLFGSSDD311 pKa = 3.57DD312 pKa = 4.36GIEE315 pKa = 4.15LFGGTVDD322 pKa = 3.05IKK324 pKa = 11.11YY325 pKa = 10.12LVVYY329 pKa = 10.02GADD332 pKa = 4.11DD333 pKa = 4.4DD334 pKa = 5.52SFDD337 pKa = 4.69YY338 pKa = 11.19DD339 pKa = 3.78QGWSGKK345 pKa = 7.89GQFWLAVQRR354 pKa = 11.84DD355 pKa = 3.84GADD358 pKa = 2.9QGFEE362 pKa = 3.97NDD364 pKa = 3.56GCADD368 pKa = 4.62EE369 pKa = 5.55ADD371 pKa = 4.47CDD373 pKa = 3.82QGSGPGSPNISNATVIAPKK392 pKa = 9.15NTASGKK398 pKa = 10.64DD399 pKa = 3.45NMGLKK404 pKa = 10.17LRR406 pKa = 11.84EE407 pKa = 4.27GLQGAYY413 pKa = 10.46RR414 pKa = 11.84NIIVSNFVGNPFYY427 pKa = 11.2LDD429 pKa = 3.61GEE431 pKa = 4.48NNANDD436 pKa = 3.97DD437 pKa = 3.7TYY439 pKa = 11.79EE440 pKa = 4.42NYY442 pKa = 10.64DD443 pKa = 3.34SGDD446 pKa = 3.41LVLEE450 pKa = 4.15NFVLQNNGPWASSDD464 pKa = 3.16EE465 pKa = 4.23SRR467 pKa = 11.84YY468 pKa = 9.47SAVYY472 pKa = 9.98EE473 pKa = 4.11YY474 pKa = 10.76TSSDD478 pKa = 3.24KK479 pKa = 11.09PADD482 pKa = 3.71LFNNPSNYY490 pKa = 10.11DD491 pKa = 3.24YY492 pKa = 11.34SLPSGSAYY500 pKa = 10.66LSGGQAPSDD509 pKa = 3.31SWFDD513 pKa = 3.54DD514 pKa = 3.39VSFRR518 pKa = 11.84GAVGEE523 pKa = 6.14SGTQWDD529 pKa = 4.3WVNEE533 pKa = 4.69GIWVKK538 pKa = 10.39WPII541 pKa = 3.56
Molecular weight: 57.35 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M1TIF2|A0A6M1TIF2_9BACT UvrABC system protein C OS=Aliifodinibius halophilus OX=1736908 GN=uvrC PE=3 SV=1
PP1 pKa = 7.45AAEE4 pKa = 3.87QSLRR8 pKa = 11.84QCLEE12 pKa = 3.78TLGQQIDD19 pKa = 3.52QLTRR23 pKa = 11.84LIRR26 pKa = 11.84KK27 pKa = 8.09HH28 pKa = 5.26LRR30 pKa = 11.84SRR32 pKa = 11.84EE33 pKa = 3.89EE34 pKa = 3.52LRR36 pKa = 11.84EE37 pKa = 4.09SIKK40 pKa = 10.68YY41 pKa = 10.11LSSIPGIGILTIAVVLAEE59 pKa = 4.24TTGFQQFHH67 pKa = 7.48KK68 pKa = 9.89ISQLISFSGYY78 pKa = 10.63DD79 pKa = 3.28VIIRR83 pKa = 11.84QSGKK87 pKa = 8.45WAGKK91 pKa = 9.25PRR93 pKa = 11.84ISKK96 pKa = 10.19QGSKK100 pKa = 10.49YY101 pKa = 9.6IRR103 pKa = 11.84RR104 pKa = 11.84AMFMPASAVVRR115 pKa = 11.84SGTGPTYY122 pKa = 10.79RR123 pKa = 11.84LYY125 pKa = 11.21
PP1 pKa = 7.45AAEE4 pKa = 3.87QSLRR8 pKa = 11.84QCLEE12 pKa = 3.78TLGQQIDD19 pKa = 3.52QLTRR23 pKa = 11.84LIRR26 pKa = 11.84KK27 pKa = 8.09HH28 pKa = 5.26LRR30 pKa = 11.84SRR32 pKa = 11.84EE33 pKa = 3.89EE34 pKa = 3.52LRR36 pKa = 11.84EE37 pKa = 4.09SIKK40 pKa = 10.68YY41 pKa = 10.11LSSIPGIGILTIAVVLAEE59 pKa = 4.24TTGFQQFHH67 pKa = 7.48KK68 pKa = 9.89ISQLISFSGYY78 pKa = 10.63DD79 pKa = 3.28VIIRR83 pKa = 11.84QSGKK87 pKa = 8.45WAGKK91 pKa = 9.25PRR93 pKa = 11.84ISKK96 pKa = 10.19QGSKK100 pKa = 10.49YY101 pKa = 9.6IRR103 pKa = 11.84RR104 pKa = 11.84AMFMPASAVVRR115 pKa = 11.84SGTGPTYY122 pKa = 10.79RR123 pKa = 11.84LYY125 pKa = 11.21
Molecular weight: 14.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1192668 |
21 |
3099 |
332.2 |
37.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.742 ± 0.041 | 0.667 ± 0.012 |
6.138 ± 0.039 | 7.352 ± 0.052 |
4.493 ± 0.031 | 6.911 ± 0.043 |
2.007 ± 0.019 | 6.978 ± 0.039 |
6.248 ± 0.042 | 9.192 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.314 ± 0.024 | 4.966 ± 0.042 |
3.866 ± 0.024 | 4.047 ± 0.027 |
4.39 ± 0.034 | 6.835 ± 0.044 |
5.648 ± 0.046 | 6.252 ± 0.03 |
1.236 ± 0.016 | 3.718 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
