
Cedar virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Henipavirus; Cedar henipavirus
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
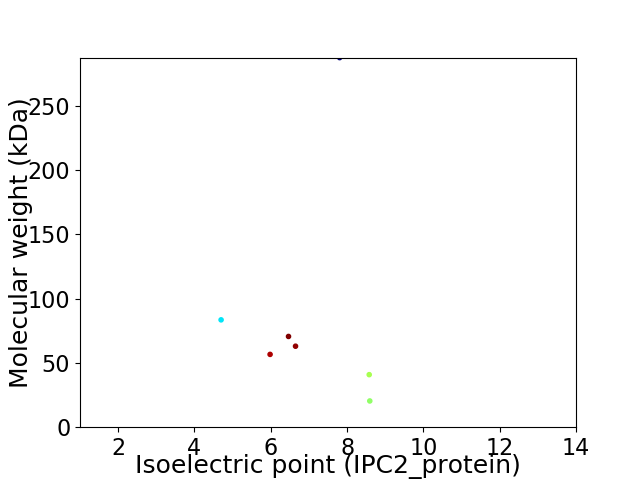
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
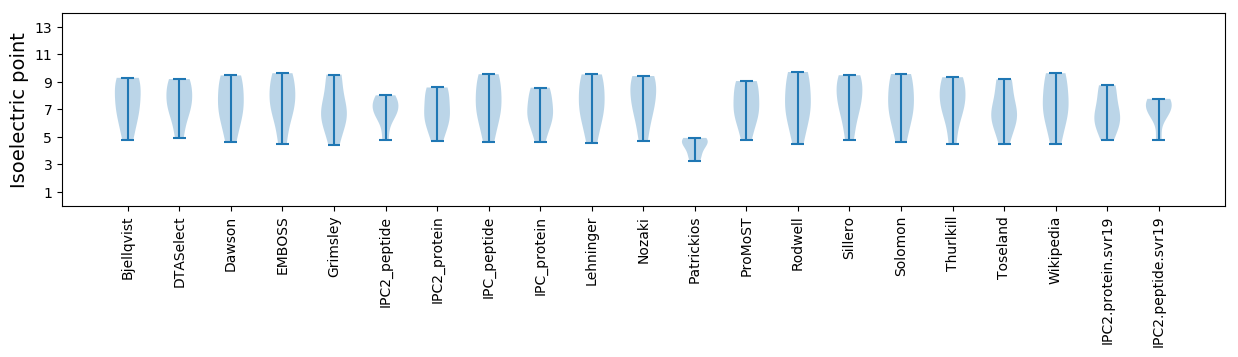
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J7GXK9|J7GXK9_9MONO GDP polyribonucleotidyltransferase OS=Cedar virus OX=1221391 GN=L PE=3 SV=1
MM1 pKa = 8.2DD2 pKa = 5.37KK3 pKa = 10.94LQLIEE8 pKa = 5.95DD9 pKa = 4.16GLSTINFIQEE19 pKa = 4.0NKK21 pKa = 10.16EE22 pKa = 4.07KK23 pKa = 10.43LQHH26 pKa = 5.88SYY28 pKa = 10.56GRR30 pKa = 11.84SSIRR34 pKa = 11.84EE35 pKa = 3.97PPTSVRR41 pKa = 11.84VEE43 pKa = 3.82EE44 pKa = 4.17WEE46 pKa = 3.95KK47 pKa = 10.72FIRR50 pKa = 11.84KK51 pKa = 8.52IASGPEE57 pKa = 3.85QVQGGGSEE65 pKa = 4.28TEE67 pKa = 3.77ITGDD71 pKa = 3.21NGDD74 pKa = 4.06RR75 pKa = 11.84GNFTNPDD82 pKa = 3.05QGGGVTGQFEE92 pKa = 4.1EE93 pKa = 6.24RR94 pKa = 11.84YY95 pKa = 9.42QKK97 pKa = 10.02WGSQDD102 pKa = 3.96SEE104 pKa = 4.43LQLDD108 pKa = 3.88PMVVHH113 pKa = 7.27DD114 pKa = 4.41FFYY117 pKa = 10.91DD118 pKa = 3.6EE119 pKa = 5.0RR120 pKa = 11.84RR121 pKa = 11.84EE122 pKa = 3.94NPDD125 pKa = 2.74NGKK128 pKa = 10.11YY129 pKa = 10.09DD130 pKa = 3.63RR131 pKa = 11.84SSKK134 pKa = 10.47KK135 pKa = 9.54RR136 pKa = 11.84DD137 pKa = 3.57NIRR140 pKa = 11.84EE141 pKa = 4.05GTRR144 pKa = 11.84QDD146 pKa = 3.19KK147 pKa = 11.01YY148 pKa = 11.32NNQSTDD154 pKa = 3.16EE155 pKa = 4.7LLSCLQPSSKK165 pKa = 10.95NDD167 pKa = 3.36VIKK170 pKa = 11.14NEE172 pKa = 4.17STSVSNLHH180 pKa = 5.11VTGNKK185 pKa = 9.63LNPDD189 pKa = 4.05AKK191 pKa = 10.18PFEE194 pKa = 4.46PTSQSKK200 pKa = 7.81EE201 pKa = 4.08HH202 pKa = 6.63PTTTQHH208 pKa = 6.35NKK210 pKa = 9.71NDD212 pKa = 3.54HH213 pKa = 5.47QTDD216 pKa = 3.41DD217 pKa = 3.73DD218 pKa = 4.55YY219 pKa = 12.04KK220 pKa = 11.08NRR222 pKa = 11.84RR223 pKa = 11.84SSEE226 pKa = 3.89NNVISDD232 pKa = 3.79HH233 pKa = 5.79ATTMEE238 pKa = 4.83DD239 pKa = 3.19NNNFIPATKK248 pKa = 9.99RR249 pKa = 11.84KK250 pKa = 9.02NALSEE255 pKa = 4.36PIYY258 pKa = 10.7VQVLPSNTEE267 pKa = 3.88GFSGKK272 pKa = 10.03DD273 pKa = 3.36YY274 pKa = 11.2PLLKK278 pKa = 10.75DD279 pKa = 3.4NSVKK283 pKa = 10.2KK284 pKa = 10.19RR285 pKa = 11.84AEE287 pKa = 3.83PVILEE292 pKa = 4.5TANHH296 pKa = 6.67PAGSADD302 pKa = 3.22QDD304 pKa = 3.86TNQIEE309 pKa = 4.22EE310 pKa = 4.14NMQFNLPKK318 pKa = 10.68LLTEE322 pKa = 4.67DD323 pKa = 3.91TDD325 pKa = 6.4DD326 pKa = 4.5EE327 pKa = 5.16PEE329 pKa = 4.55DD330 pKa = 4.68NNDD333 pKa = 3.58SMPLEE338 pKa = 4.02EE339 pKa = 6.31DD340 pKa = 2.77IRR342 pKa = 11.84EE343 pKa = 4.13IGSMLKK349 pKa = 10.72DD350 pKa = 3.24GTKK353 pKa = 10.24DD354 pKa = 2.45IKK356 pKa = 10.37TRR358 pKa = 11.84MNEE361 pKa = 3.26IDD363 pKa = 4.36DD364 pKa = 5.01AIKK367 pKa = 10.53KK368 pKa = 9.49INKK371 pKa = 8.66KK372 pKa = 10.02SKK374 pKa = 10.14NRR376 pKa = 11.84SLDD379 pKa = 3.53LEE381 pKa = 4.54SDD383 pKa = 3.55GKK385 pKa = 10.72DD386 pKa = 3.05QGRR389 pKa = 11.84RR390 pKa = 11.84DD391 pKa = 3.65PSVDD395 pKa = 3.35LGIKK399 pKa = 9.14KK400 pKa = 10.2RR401 pKa = 11.84KK402 pKa = 9.02EE403 pKa = 3.75GLKK406 pKa = 10.61AAMQKK411 pKa = 8.91TKK413 pKa = 9.98EE414 pKa = 3.87QLSIKK419 pKa = 8.78VEE421 pKa = 4.07RR422 pKa = 11.84EE423 pKa = 3.01IGLNDD428 pKa = 5.35RR429 pKa = 11.84ICQNSKK435 pKa = 9.84MSTEE439 pKa = 3.82KK440 pKa = 10.86KK441 pKa = 10.16LIYY444 pKa = 10.37AGMEE448 pKa = 3.75MEE450 pKa = 4.61YY451 pKa = 10.7GQTSTGSGGPQGSKK465 pKa = 10.68DD466 pKa = 3.52GTSDD470 pKa = 3.83DD471 pKa = 3.9VQVDD475 pKa = 3.36EE476 pKa = 6.32DD477 pKa = 4.26YY478 pKa = 11.69DD479 pKa = 3.7EE480 pKa = 6.58GEE482 pKa = 5.16DD483 pKa = 3.93YY484 pKa = 11.37EE485 pKa = 5.59AMPSDD490 pKa = 3.59RR491 pKa = 11.84FYY493 pKa = 9.19TTLSGEE499 pKa = 4.07QKK501 pKa = 10.94DD502 pKa = 5.22RR503 pKa = 11.84FDD505 pKa = 5.02LDD507 pKa = 3.84ANQMSQYY514 pKa = 10.98DD515 pKa = 3.88LEE517 pKa = 4.53AQVDD521 pKa = 3.96EE522 pKa = 4.5LTRR525 pKa = 11.84MNLILYY531 pKa = 9.99SRR533 pKa = 11.84LEE535 pKa = 4.18TTNKK539 pKa = 10.32LLIDD543 pKa = 3.73ILDD546 pKa = 4.21LAKK549 pKa = 10.63EE550 pKa = 4.05MPKK553 pKa = 10.39LVRR556 pKa = 11.84KK557 pKa = 9.78VDD559 pKa = 3.45NLEE562 pKa = 3.94RR563 pKa = 11.84QMGNLNMLTSTLEE576 pKa = 3.94GHH578 pKa = 7.15LSSVMIMIPGKK589 pKa = 10.2DD590 pKa = 3.28KK591 pKa = 11.34SEE593 pKa = 4.24KK594 pKa = 10.09EE595 pKa = 3.83IPKK598 pKa = 10.6NPDD601 pKa = 2.73LRR603 pKa = 11.84PILGRR608 pKa = 11.84SNTSLTDD615 pKa = 3.84VIDD618 pKa = 4.36LDD620 pKa = 4.11HH621 pKa = 7.0YY622 pKa = 10.19PDD624 pKa = 4.71KK625 pKa = 10.85GSKK628 pKa = 9.89GIKK631 pKa = 9.44PSGSGDD637 pKa = 3.2RR638 pKa = 11.84QYY640 pKa = 11.15IGSLEE645 pKa = 4.39SKK647 pKa = 10.5FSINDD652 pKa = 3.3EE653 pKa = 4.63YY654 pKa = 11.81NFAPYY659 pKa = 9.81PIRR662 pKa = 11.84DD663 pKa = 3.74EE664 pKa = 4.51LLLPGLRR671 pKa = 11.84DD672 pKa = 3.73DD673 pKa = 4.36KK674 pKa = 11.35TNASSFIPDD683 pKa = 3.49DD684 pKa = 3.65TDD686 pKa = 3.36RR687 pKa = 11.84SPMVLKK693 pKa = 10.59IIIRR697 pKa = 11.84QNIHH701 pKa = 6.49DD702 pKa = 4.51EE703 pKa = 4.22EE704 pKa = 4.74VKK706 pKa = 11.09DD707 pKa = 3.76EE708 pKa = 4.38LLSILEE714 pKa = 3.83QHH716 pKa = 5.77NTVEE720 pKa = 4.35EE721 pKa = 4.41LNEE724 pKa = 3.51IWNTVNDD731 pKa = 4.05YY732 pKa = 11.56LDD734 pKa = 4.0GNII737 pKa = 4.56
MM1 pKa = 8.2DD2 pKa = 5.37KK3 pKa = 10.94LQLIEE8 pKa = 5.95DD9 pKa = 4.16GLSTINFIQEE19 pKa = 4.0NKK21 pKa = 10.16EE22 pKa = 4.07KK23 pKa = 10.43LQHH26 pKa = 5.88SYY28 pKa = 10.56GRR30 pKa = 11.84SSIRR34 pKa = 11.84EE35 pKa = 3.97PPTSVRR41 pKa = 11.84VEE43 pKa = 3.82EE44 pKa = 4.17WEE46 pKa = 3.95KK47 pKa = 10.72FIRR50 pKa = 11.84KK51 pKa = 8.52IASGPEE57 pKa = 3.85QVQGGGSEE65 pKa = 4.28TEE67 pKa = 3.77ITGDD71 pKa = 3.21NGDD74 pKa = 4.06RR75 pKa = 11.84GNFTNPDD82 pKa = 3.05QGGGVTGQFEE92 pKa = 4.1EE93 pKa = 6.24RR94 pKa = 11.84YY95 pKa = 9.42QKK97 pKa = 10.02WGSQDD102 pKa = 3.96SEE104 pKa = 4.43LQLDD108 pKa = 3.88PMVVHH113 pKa = 7.27DD114 pKa = 4.41FFYY117 pKa = 10.91DD118 pKa = 3.6EE119 pKa = 5.0RR120 pKa = 11.84RR121 pKa = 11.84EE122 pKa = 3.94NPDD125 pKa = 2.74NGKK128 pKa = 10.11YY129 pKa = 10.09DD130 pKa = 3.63RR131 pKa = 11.84SSKK134 pKa = 10.47KK135 pKa = 9.54RR136 pKa = 11.84DD137 pKa = 3.57NIRR140 pKa = 11.84EE141 pKa = 4.05GTRR144 pKa = 11.84QDD146 pKa = 3.19KK147 pKa = 11.01YY148 pKa = 11.32NNQSTDD154 pKa = 3.16EE155 pKa = 4.7LLSCLQPSSKK165 pKa = 10.95NDD167 pKa = 3.36VIKK170 pKa = 11.14NEE172 pKa = 4.17STSVSNLHH180 pKa = 5.11VTGNKK185 pKa = 9.63LNPDD189 pKa = 4.05AKK191 pKa = 10.18PFEE194 pKa = 4.46PTSQSKK200 pKa = 7.81EE201 pKa = 4.08HH202 pKa = 6.63PTTTQHH208 pKa = 6.35NKK210 pKa = 9.71NDD212 pKa = 3.54HH213 pKa = 5.47QTDD216 pKa = 3.41DD217 pKa = 3.73DD218 pKa = 4.55YY219 pKa = 12.04KK220 pKa = 11.08NRR222 pKa = 11.84RR223 pKa = 11.84SSEE226 pKa = 3.89NNVISDD232 pKa = 3.79HH233 pKa = 5.79ATTMEE238 pKa = 4.83DD239 pKa = 3.19NNNFIPATKK248 pKa = 9.99RR249 pKa = 11.84KK250 pKa = 9.02NALSEE255 pKa = 4.36PIYY258 pKa = 10.7VQVLPSNTEE267 pKa = 3.88GFSGKK272 pKa = 10.03DD273 pKa = 3.36YY274 pKa = 11.2PLLKK278 pKa = 10.75DD279 pKa = 3.4NSVKK283 pKa = 10.2KK284 pKa = 10.19RR285 pKa = 11.84AEE287 pKa = 3.83PVILEE292 pKa = 4.5TANHH296 pKa = 6.67PAGSADD302 pKa = 3.22QDD304 pKa = 3.86TNQIEE309 pKa = 4.22EE310 pKa = 4.14NMQFNLPKK318 pKa = 10.68LLTEE322 pKa = 4.67DD323 pKa = 3.91TDD325 pKa = 6.4DD326 pKa = 4.5EE327 pKa = 5.16PEE329 pKa = 4.55DD330 pKa = 4.68NNDD333 pKa = 3.58SMPLEE338 pKa = 4.02EE339 pKa = 6.31DD340 pKa = 2.77IRR342 pKa = 11.84EE343 pKa = 4.13IGSMLKK349 pKa = 10.72DD350 pKa = 3.24GTKK353 pKa = 10.24DD354 pKa = 2.45IKK356 pKa = 10.37TRR358 pKa = 11.84MNEE361 pKa = 3.26IDD363 pKa = 4.36DD364 pKa = 5.01AIKK367 pKa = 10.53KK368 pKa = 9.49INKK371 pKa = 8.66KK372 pKa = 10.02SKK374 pKa = 10.14NRR376 pKa = 11.84SLDD379 pKa = 3.53LEE381 pKa = 4.54SDD383 pKa = 3.55GKK385 pKa = 10.72DD386 pKa = 3.05QGRR389 pKa = 11.84RR390 pKa = 11.84DD391 pKa = 3.65PSVDD395 pKa = 3.35LGIKK399 pKa = 9.14KK400 pKa = 10.2RR401 pKa = 11.84KK402 pKa = 9.02EE403 pKa = 3.75GLKK406 pKa = 10.61AAMQKK411 pKa = 8.91TKK413 pKa = 9.98EE414 pKa = 3.87QLSIKK419 pKa = 8.78VEE421 pKa = 4.07RR422 pKa = 11.84EE423 pKa = 3.01IGLNDD428 pKa = 5.35RR429 pKa = 11.84ICQNSKK435 pKa = 9.84MSTEE439 pKa = 3.82KK440 pKa = 10.86KK441 pKa = 10.16LIYY444 pKa = 10.37AGMEE448 pKa = 3.75MEE450 pKa = 4.61YY451 pKa = 10.7GQTSTGSGGPQGSKK465 pKa = 10.68DD466 pKa = 3.52GTSDD470 pKa = 3.83DD471 pKa = 3.9VQVDD475 pKa = 3.36EE476 pKa = 6.32DD477 pKa = 4.26YY478 pKa = 11.69DD479 pKa = 3.7EE480 pKa = 6.58GEE482 pKa = 5.16DD483 pKa = 3.93YY484 pKa = 11.37EE485 pKa = 5.59AMPSDD490 pKa = 3.59RR491 pKa = 11.84FYY493 pKa = 9.19TTLSGEE499 pKa = 4.07QKK501 pKa = 10.94DD502 pKa = 5.22RR503 pKa = 11.84FDD505 pKa = 5.02LDD507 pKa = 3.84ANQMSQYY514 pKa = 10.98DD515 pKa = 3.88LEE517 pKa = 4.53AQVDD521 pKa = 3.96EE522 pKa = 4.5LTRR525 pKa = 11.84MNLILYY531 pKa = 9.99SRR533 pKa = 11.84LEE535 pKa = 4.18TTNKK539 pKa = 10.32LLIDD543 pKa = 3.73ILDD546 pKa = 4.21LAKK549 pKa = 10.63EE550 pKa = 4.05MPKK553 pKa = 10.39LVRR556 pKa = 11.84KK557 pKa = 9.78VDD559 pKa = 3.45NLEE562 pKa = 3.94RR563 pKa = 11.84QMGNLNMLTSTLEE576 pKa = 3.94GHH578 pKa = 7.15LSSVMIMIPGKK589 pKa = 10.2DD590 pKa = 3.28KK591 pKa = 11.34SEE593 pKa = 4.24KK594 pKa = 10.09EE595 pKa = 3.83IPKK598 pKa = 10.6NPDD601 pKa = 2.73LRR603 pKa = 11.84PILGRR608 pKa = 11.84SNTSLTDD615 pKa = 3.84VIDD618 pKa = 4.36LDD620 pKa = 4.11HH621 pKa = 7.0YY622 pKa = 10.19PDD624 pKa = 4.71KK625 pKa = 10.85GSKK628 pKa = 9.89GIKK631 pKa = 9.44PSGSGDD637 pKa = 3.2RR638 pKa = 11.84QYY640 pKa = 11.15IGSLEE645 pKa = 4.39SKK647 pKa = 10.5FSINDD652 pKa = 3.3EE653 pKa = 4.63YY654 pKa = 11.81NFAPYY659 pKa = 9.81PIRR662 pKa = 11.84DD663 pKa = 3.74EE664 pKa = 4.51LLLPGLRR671 pKa = 11.84DD672 pKa = 3.73DD673 pKa = 4.36KK674 pKa = 11.35TNASSFIPDD683 pKa = 3.49DD684 pKa = 3.65TDD686 pKa = 3.36RR687 pKa = 11.84SPMVLKK693 pKa = 10.59IIIRR697 pKa = 11.84QNIHH701 pKa = 6.49DD702 pKa = 4.51EE703 pKa = 4.22EE704 pKa = 4.74VKK706 pKa = 11.09DD707 pKa = 3.76EE708 pKa = 4.38LLSILEE714 pKa = 3.83QHH716 pKa = 5.77NTVEE720 pKa = 4.35EE721 pKa = 4.41LNEE724 pKa = 3.51IWNTVNDD731 pKa = 4.05YY732 pKa = 11.56LDD734 pKa = 4.0GNII737 pKa = 4.56
Molecular weight: 83.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J7H4I1|J7H4I1_9MONO Isoform of J7GXK5 Nonstructural protein C OS=Cedar virus OX=1221391 GN=P PE=2 SV=1
MM1 pKa = 7.74ASLLSILYY9 pKa = 9.99RR10 pKa = 11.84KK11 pKa = 8.81IRR13 pKa = 11.84KK14 pKa = 9.23NYY16 pKa = 10.27SILTEE21 pKa = 4.45DD22 pKa = 4.81PPSEE26 pKa = 4.05SHH28 pKa = 6.4PQVSGLKK35 pKa = 9.7SGRR38 pKa = 11.84NLFEE42 pKa = 5.72RR43 pKa = 11.84SLLDD47 pKa = 3.59LNKK50 pKa = 10.31FKK52 pKa = 11.51GEE54 pKa = 3.98DD55 pKa = 3.28LRR57 pKa = 11.84LRR59 pKa = 11.84SQAIMEE65 pKa = 4.41IEE67 pKa = 5.06AILPILIRR75 pKa = 11.84EE76 pKa = 4.63AEE78 pKa = 4.19SQDD81 pKa = 3.35NSKK84 pKa = 10.9KK85 pKa = 9.92GIKK88 pKa = 10.08NGGHH92 pKa = 7.5KK93 pKa = 9.29IQNYY97 pKa = 8.9NWTQWLYY104 pKa = 9.98TISSMTRR111 pKa = 11.84EE112 pKa = 3.87GRR114 pKa = 11.84IPTMEE119 pKa = 4.31NMTAALKK126 pKa = 10.86NGIISEE132 pKa = 4.36KK133 pKa = 9.08EE134 pKa = 3.32HH135 pKa = 7.04DD136 pKa = 4.84RR137 pKa = 11.84ISTIISLLMNYY148 pKa = 9.33CPAYY152 pKa = 9.9NHH154 pKa = 7.11LLRR157 pKa = 11.84TMSSRR162 pKa = 11.84MKK164 pKa = 10.11VHH166 pKa = 5.82QCQICMLQEE175 pKa = 3.94INN177 pKa = 4.08
MM1 pKa = 7.74ASLLSILYY9 pKa = 9.99RR10 pKa = 11.84KK11 pKa = 8.81IRR13 pKa = 11.84KK14 pKa = 9.23NYY16 pKa = 10.27SILTEE21 pKa = 4.45DD22 pKa = 4.81PPSEE26 pKa = 4.05SHH28 pKa = 6.4PQVSGLKK35 pKa = 9.7SGRR38 pKa = 11.84NLFEE42 pKa = 5.72RR43 pKa = 11.84SLLDD47 pKa = 3.59LNKK50 pKa = 10.31FKK52 pKa = 11.51GEE54 pKa = 3.98DD55 pKa = 3.28LRR57 pKa = 11.84LRR59 pKa = 11.84SQAIMEE65 pKa = 4.41IEE67 pKa = 5.06AILPILIRR75 pKa = 11.84EE76 pKa = 4.63AEE78 pKa = 4.19SQDD81 pKa = 3.35NSKK84 pKa = 10.9KK85 pKa = 9.92GIKK88 pKa = 10.08NGGHH92 pKa = 7.5KK93 pKa = 9.29IQNYY97 pKa = 8.9NWTQWLYY104 pKa = 9.98TISSMTRR111 pKa = 11.84EE112 pKa = 3.87GRR114 pKa = 11.84IPTMEE119 pKa = 4.31NMTAALKK126 pKa = 10.86NGIISEE132 pKa = 4.36KK133 pKa = 9.08EE134 pKa = 3.32HH135 pKa = 7.04DD136 pKa = 4.84RR137 pKa = 11.84ISTIISLLMNYY148 pKa = 9.33CPAYY152 pKa = 9.9NHH154 pKa = 7.11LLRR157 pKa = 11.84TMSSRR162 pKa = 11.84MKK164 pKa = 10.11VHH166 pKa = 5.82QCQICMLQEE175 pKa = 3.94INN177 pKa = 4.08
Molecular weight: 20.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5464 |
177 |
2501 |
780.6 |
88.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.734 ± 0.695 | 1.574 ± 0.407 |
6.57 ± 0.667 | 5.783 ± 0.641 |
3.715 ± 0.445 | 5.179 ± 0.425 |
2.05 ± 0.464 | 8.547 ± 0.724 |
7.485 ± 0.318 | 9.023 ± 0.28 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.928 ± 0.328 | 7.174 ± 0.533 |
4.228 ± 0.188 | 3.459 ± 0.33 |
4.85 ± 0.46 | 8.272 ± 0.491 |
5.966 ± 0.222 | 4.758 ± 0.416 |
0.86 ± 0.19 | 3.843 ± 0.465 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
