
Novosphingobium aromaticivorans (strain ATCC 700278 / DSM 12444 / CCUG 56034 / CIP 105152 / NBRC 16084 / F199)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium; Novosphingobium aromaticivorans
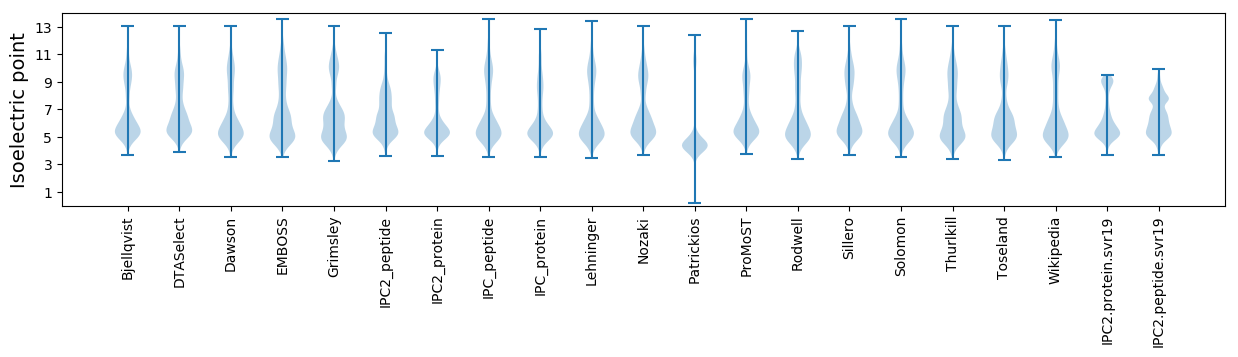
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
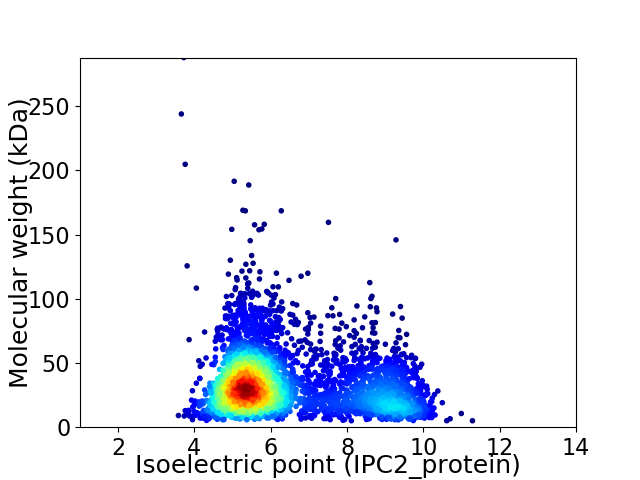
Virtual 2D-PAGE plot for 3917 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q2G7Y7|Q2G7Y7_NOVAD Type IV secretory pathway VirJ component-like protein OS=Novosphingobium aromaticivorans (strain ATCC 700278 / DSM 12444 / CCUG 56034 / CIP 105152 / NBRC 16084 / F199) OX=279238 GN=Saro_1596 PE=4 SV=1
MM1 pKa = 7.73TYY3 pKa = 10.17IQIDD7 pKa = 3.93GSLSDD12 pKa = 3.16WSSNLRR18 pKa = 11.84IDD20 pKa = 3.57AGAVDD25 pKa = 5.59GYY27 pKa = 10.82QIYY30 pKa = 8.16ATTDD34 pKa = 2.7ATDD37 pKa = 3.59YY38 pKa = 10.68IFAFAAPTAVGANTTIWLNTDD59 pKa = 3.97LNQATGYY66 pKa = 8.41QLWGTVGAEE75 pKa = 3.67FNVNFKK81 pKa = 11.01SDD83 pKa = 3.28GSAALYY89 pKa = 10.58SGAAGGTLVADD100 pKa = 4.11NLVLAYY106 pKa = 10.55NADD109 pKa = 3.26KK110 pKa = 11.27TMVEE114 pKa = 3.9LRR116 pKa = 11.84VPKK119 pKa = 10.37DD120 pKa = 3.17LLGNPGSIDD129 pKa = 3.25TVYY132 pKa = 10.73DD133 pKa = 3.49INDD136 pKa = 3.58TAIIPSFYY144 pKa = 9.67QDD146 pKa = 2.75NAIRR150 pKa = 11.84VWDD153 pKa = 4.28DD154 pKa = 3.47SEE156 pKa = 4.19LASVIPATDD165 pKa = 3.06TRR167 pKa = 11.84IAIVYY172 pKa = 9.57SATTAANYY180 pKa = 9.34FSQTAYY186 pKa = 11.1SDD188 pKa = 3.91LFMAAQSQAAQAGVPFDD205 pKa = 4.93IITEE209 pKa = 4.12ADD211 pKa = 3.25LTDD214 pKa = 3.57INKK217 pKa = 9.48LAQYY221 pKa = 9.69KK222 pKa = 10.62AIVFPSFRR230 pKa = 11.84NVQASQADD238 pKa = 4.58EE239 pKa = 4.11IAHH242 pKa = 5.93TLQLASQEE250 pKa = 4.06FHH252 pKa = 6.59VGFIVSGEE260 pKa = 3.82FMTNDD265 pKa = 3.2EE266 pKa = 5.1NGNAMAGNSYY276 pKa = 11.15SRR278 pKa = 11.84MATLLDD284 pKa = 3.43ATRR287 pKa = 11.84VTGGTATSLTVTATDD302 pKa = 3.54PTGVVLDD309 pKa = 5.13GYY311 pKa = 11.83ANGEE315 pKa = 4.33LVNQYY320 pKa = 11.8ANVGWNAFQSVSGTGQTIATEE341 pKa = 4.62TINGSSTYY349 pKa = 10.72AAVLATQTGGRR360 pKa = 11.84NVLFSSDD367 pKa = 3.76AVMADD372 pKa = 2.83ANMLQRR378 pKa = 11.84AIDD381 pKa = 3.77YY382 pKa = 9.16AVSGEE387 pKa = 4.34TVTVSLNMTRR397 pKa = 11.84DD398 pKa = 3.35AGLVAARR405 pKa = 11.84VDD407 pKa = 4.17MDD409 pKa = 3.04QSMYY413 pKa = 10.59IEE415 pKa = 4.8DD416 pKa = 4.26VNGGIYY422 pKa = 10.18DD423 pKa = 3.51QLVPLLQQWKK433 pKa = 8.6AQYY436 pKa = 10.58NFVGSFYY443 pKa = 11.61VNIGDD448 pKa = 3.69NTQQGIYY455 pKa = 9.54TDD457 pKa = 3.72WNKK460 pKa = 10.79SLPYY464 pKa = 7.38YY465 pKa = 9.85TAMIGLGNEE474 pKa = 4.32IGTHH478 pKa = 5.99TYY480 pKa = 7.58THH482 pKa = 7.2PEE484 pKa = 4.0DD485 pKa = 4.12TNLLSPSQLQFEE497 pKa = 5.18FEE499 pKa = 4.67LSTQILEE506 pKa = 4.46QKK508 pKa = 10.55LSAALGYY515 pKa = 10.71AYY517 pKa = 9.2TIEE520 pKa = 4.38GAAIPGAPEE529 pKa = 3.76TLTTSLAIEE538 pKa = 4.42QYY540 pKa = 10.6VKK542 pKa = 10.12TYY544 pKa = 9.4LTGGYY549 pKa = 7.65TGQGAGYY556 pKa = 8.58PNAFGYY562 pKa = 7.44LTPGSQDD569 pKa = 2.87KK570 pKa = 11.23VYY572 pKa = 10.35IAPNTFFDD580 pKa = 3.82FTLFDD585 pKa = 3.94WLHH588 pKa = 6.45LSAADD593 pKa = 5.8ASALWQSQYY602 pKa = 11.47EE603 pKa = 4.5KK604 pKa = 10.34IVSQADD610 pKa = 3.66SPVVVWPWHH619 pKa = 7.13DD620 pKa = 3.77YY621 pKa = 10.79GATAFNSPNYY631 pKa = 10.21APEE634 pKa = 3.94IFNTFLAQAAADD646 pKa = 3.45GMEE649 pKa = 4.15FVTLADD655 pKa = 3.54LANRR659 pKa = 11.84INAFHH664 pKa = 6.57GAKK667 pKa = 8.86VTTSVSGNTITANVTASGNVGTFAFDD693 pKa = 3.44LQGQGSQVISSVAGWYY709 pKa = 10.19AYY711 pKa = 10.58DD712 pKa = 3.7SDD714 pKa = 5.71SVFLPQNGGTFVITLGAAQTDD735 pKa = 3.78VTHH738 pKa = 7.57IIDD741 pKa = 3.89LPMRR745 pKa = 11.84ATLMSVTGNGTNLSFQIQGEE765 pKa = 4.55GTVVIDD771 pKa = 6.01LSDD774 pKa = 3.7PTNKK778 pKa = 9.83SVQVSGATIVSQVGDD793 pKa = 4.23KK794 pKa = 10.05LTIDD798 pKa = 3.65IGPVGSHH805 pKa = 5.33TVTVTQTSLNHH816 pKa = 6.16APVIEE821 pKa = 4.53SNGGGDD827 pKa = 3.26TAAISLAEE835 pKa = 4.12NLLAVTAVIATDD847 pKa = 3.36ADD849 pKa = 4.17ANALTYY855 pKa = 10.51SITGGADD862 pKa = 2.91ASKK865 pKa = 9.22FTINATTGALAFLAAPNFEE884 pKa = 4.22VPTDD888 pKa = 3.48VGGNNVYY895 pKa = 10.74DD896 pKa = 4.03VVVTASDD903 pKa = 3.74GALTDD908 pKa = 3.8SQALAVTVTNVNEE921 pKa = 4.19APVITSNGGGATASISLAEE940 pKa = 4.28NNAAVTVVTSTDD952 pKa = 3.46PEE954 pKa = 4.18NTARR958 pKa = 11.84TYY960 pKa = 11.27SLSGTDD966 pKa = 3.19AARR969 pKa = 11.84FTIDD973 pKa = 3.17AATGALSFVNAPDD986 pKa = 3.9FEE988 pKa = 4.99NPTDD992 pKa = 3.45VGANNVYY999 pKa = 10.51NVVVTASDD1007 pKa = 3.66GSLTDD1012 pKa = 3.57TQALAITVTNKK1023 pKa = 10.46KK1024 pKa = 10.3GVTLNASSSTGSVLNGTGEE1043 pKa = 4.08EE1044 pKa = 4.09DD1045 pKa = 4.73QLNGWKK1051 pKa = 10.49GADD1054 pKa = 3.21TLYY1057 pKa = 11.48GLGGNDD1063 pKa = 3.48RR1064 pKa = 11.84LDD1066 pKa = 3.75GAGGNDD1072 pKa = 3.2RR1073 pKa = 11.84LYY1075 pKa = 11.23GGDD1078 pKa = 3.83GKK1080 pKa = 10.73DD1081 pKa = 3.51VLIGGAGTDD1090 pKa = 3.42IMSGGAGADD1099 pKa = 2.72RR1100 pKa = 11.84FEE1102 pKa = 5.04FNALGNSVTGALHH1115 pKa = 7.39DD1116 pKa = 4.53VITDD1120 pKa = 3.81FEE1122 pKa = 5.38AGIDD1126 pKa = 4.09LIDD1129 pKa = 4.14VSSIDD1134 pKa = 3.66ANSGKK1139 pKa = 10.5GGNQTFVLLAEE1150 pKa = 4.95GAAFTGVGQLRR1161 pKa = 11.84YY1162 pKa = 9.25FYY1164 pKa = 11.04DD1165 pKa = 3.41SATDD1169 pKa = 3.41QTIVQGNVNNNLAADD1184 pKa = 4.03FEE1186 pKa = 4.78IALSGHH1192 pKa = 4.87QTLSASMFILL1202 pKa = 4.18
MM1 pKa = 7.73TYY3 pKa = 10.17IQIDD7 pKa = 3.93GSLSDD12 pKa = 3.16WSSNLRR18 pKa = 11.84IDD20 pKa = 3.57AGAVDD25 pKa = 5.59GYY27 pKa = 10.82QIYY30 pKa = 8.16ATTDD34 pKa = 2.7ATDD37 pKa = 3.59YY38 pKa = 10.68IFAFAAPTAVGANTTIWLNTDD59 pKa = 3.97LNQATGYY66 pKa = 8.41QLWGTVGAEE75 pKa = 3.67FNVNFKK81 pKa = 11.01SDD83 pKa = 3.28GSAALYY89 pKa = 10.58SGAAGGTLVADD100 pKa = 4.11NLVLAYY106 pKa = 10.55NADD109 pKa = 3.26KK110 pKa = 11.27TMVEE114 pKa = 3.9LRR116 pKa = 11.84VPKK119 pKa = 10.37DD120 pKa = 3.17LLGNPGSIDD129 pKa = 3.25TVYY132 pKa = 10.73DD133 pKa = 3.49INDD136 pKa = 3.58TAIIPSFYY144 pKa = 9.67QDD146 pKa = 2.75NAIRR150 pKa = 11.84VWDD153 pKa = 4.28DD154 pKa = 3.47SEE156 pKa = 4.19LASVIPATDD165 pKa = 3.06TRR167 pKa = 11.84IAIVYY172 pKa = 9.57SATTAANYY180 pKa = 9.34FSQTAYY186 pKa = 11.1SDD188 pKa = 3.91LFMAAQSQAAQAGVPFDD205 pKa = 4.93IITEE209 pKa = 4.12ADD211 pKa = 3.25LTDD214 pKa = 3.57INKK217 pKa = 9.48LAQYY221 pKa = 9.69KK222 pKa = 10.62AIVFPSFRR230 pKa = 11.84NVQASQADD238 pKa = 4.58EE239 pKa = 4.11IAHH242 pKa = 5.93TLQLASQEE250 pKa = 4.06FHH252 pKa = 6.59VGFIVSGEE260 pKa = 3.82FMTNDD265 pKa = 3.2EE266 pKa = 5.1NGNAMAGNSYY276 pKa = 11.15SRR278 pKa = 11.84MATLLDD284 pKa = 3.43ATRR287 pKa = 11.84VTGGTATSLTVTATDD302 pKa = 3.54PTGVVLDD309 pKa = 5.13GYY311 pKa = 11.83ANGEE315 pKa = 4.33LVNQYY320 pKa = 11.8ANVGWNAFQSVSGTGQTIATEE341 pKa = 4.62TINGSSTYY349 pKa = 10.72AAVLATQTGGRR360 pKa = 11.84NVLFSSDD367 pKa = 3.76AVMADD372 pKa = 2.83ANMLQRR378 pKa = 11.84AIDD381 pKa = 3.77YY382 pKa = 9.16AVSGEE387 pKa = 4.34TVTVSLNMTRR397 pKa = 11.84DD398 pKa = 3.35AGLVAARR405 pKa = 11.84VDD407 pKa = 4.17MDD409 pKa = 3.04QSMYY413 pKa = 10.59IEE415 pKa = 4.8DD416 pKa = 4.26VNGGIYY422 pKa = 10.18DD423 pKa = 3.51QLVPLLQQWKK433 pKa = 8.6AQYY436 pKa = 10.58NFVGSFYY443 pKa = 11.61VNIGDD448 pKa = 3.69NTQQGIYY455 pKa = 9.54TDD457 pKa = 3.72WNKK460 pKa = 10.79SLPYY464 pKa = 7.38YY465 pKa = 9.85TAMIGLGNEE474 pKa = 4.32IGTHH478 pKa = 5.99TYY480 pKa = 7.58THH482 pKa = 7.2PEE484 pKa = 4.0DD485 pKa = 4.12TNLLSPSQLQFEE497 pKa = 5.18FEE499 pKa = 4.67LSTQILEE506 pKa = 4.46QKK508 pKa = 10.55LSAALGYY515 pKa = 10.71AYY517 pKa = 9.2TIEE520 pKa = 4.38GAAIPGAPEE529 pKa = 3.76TLTTSLAIEE538 pKa = 4.42QYY540 pKa = 10.6VKK542 pKa = 10.12TYY544 pKa = 9.4LTGGYY549 pKa = 7.65TGQGAGYY556 pKa = 8.58PNAFGYY562 pKa = 7.44LTPGSQDD569 pKa = 2.87KK570 pKa = 11.23VYY572 pKa = 10.35IAPNTFFDD580 pKa = 3.82FTLFDD585 pKa = 3.94WLHH588 pKa = 6.45LSAADD593 pKa = 5.8ASALWQSQYY602 pKa = 11.47EE603 pKa = 4.5KK604 pKa = 10.34IVSQADD610 pKa = 3.66SPVVVWPWHH619 pKa = 7.13DD620 pKa = 3.77YY621 pKa = 10.79GATAFNSPNYY631 pKa = 10.21APEE634 pKa = 3.94IFNTFLAQAAADD646 pKa = 3.45GMEE649 pKa = 4.15FVTLADD655 pKa = 3.54LANRR659 pKa = 11.84INAFHH664 pKa = 6.57GAKK667 pKa = 8.86VTTSVSGNTITANVTASGNVGTFAFDD693 pKa = 3.44LQGQGSQVISSVAGWYY709 pKa = 10.19AYY711 pKa = 10.58DD712 pKa = 3.7SDD714 pKa = 5.71SVFLPQNGGTFVITLGAAQTDD735 pKa = 3.78VTHH738 pKa = 7.57IIDD741 pKa = 3.89LPMRR745 pKa = 11.84ATLMSVTGNGTNLSFQIQGEE765 pKa = 4.55GTVVIDD771 pKa = 6.01LSDD774 pKa = 3.7PTNKK778 pKa = 9.83SVQVSGATIVSQVGDD793 pKa = 4.23KK794 pKa = 10.05LTIDD798 pKa = 3.65IGPVGSHH805 pKa = 5.33TVTVTQTSLNHH816 pKa = 6.16APVIEE821 pKa = 4.53SNGGGDD827 pKa = 3.26TAAISLAEE835 pKa = 4.12NLLAVTAVIATDD847 pKa = 3.36ADD849 pKa = 4.17ANALTYY855 pKa = 10.51SITGGADD862 pKa = 2.91ASKK865 pKa = 9.22FTINATTGALAFLAAPNFEE884 pKa = 4.22VPTDD888 pKa = 3.48VGGNNVYY895 pKa = 10.74DD896 pKa = 4.03VVVTASDD903 pKa = 3.74GALTDD908 pKa = 3.8SQALAVTVTNVNEE921 pKa = 4.19APVITSNGGGATASISLAEE940 pKa = 4.28NNAAVTVVTSTDD952 pKa = 3.46PEE954 pKa = 4.18NTARR958 pKa = 11.84TYY960 pKa = 11.27SLSGTDD966 pKa = 3.19AARR969 pKa = 11.84FTIDD973 pKa = 3.17AATGALSFVNAPDD986 pKa = 3.9FEE988 pKa = 4.99NPTDD992 pKa = 3.45VGANNVYY999 pKa = 10.51NVVVTASDD1007 pKa = 3.66GSLTDD1012 pKa = 3.57TQALAITVTNKK1023 pKa = 10.46KK1024 pKa = 10.3GVTLNASSSTGSVLNGTGEE1043 pKa = 4.08EE1044 pKa = 4.09DD1045 pKa = 4.73QLNGWKK1051 pKa = 10.49GADD1054 pKa = 3.21TLYY1057 pKa = 11.48GLGGNDD1063 pKa = 3.48RR1064 pKa = 11.84LDD1066 pKa = 3.75GAGGNDD1072 pKa = 3.2RR1073 pKa = 11.84LYY1075 pKa = 11.23GGDD1078 pKa = 3.83GKK1080 pKa = 10.73DD1081 pKa = 3.51VLIGGAGTDD1090 pKa = 3.42IMSGGAGADD1099 pKa = 2.72RR1100 pKa = 11.84FEE1102 pKa = 5.04FNALGNSVTGALHH1115 pKa = 7.39DD1116 pKa = 4.53VITDD1120 pKa = 3.81FEE1122 pKa = 5.38AGIDD1126 pKa = 4.09LIDD1129 pKa = 4.14VSSIDD1134 pKa = 3.66ANSGKK1139 pKa = 10.5GGNQTFVLLAEE1150 pKa = 4.95GAAFTGVGQLRR1161 pKa = 11.84YY1162 pKa = 9.25FYY1164 pKa = 11.04DD1165 pKa = 3.41SATDD1169 pKa = 3.41QTIVQGNVNNNLAADD1184 pKa = 4.03FEE1186 pKa = 4.78IALSGHH1192 pKa = 4.87QTLSASMFILL1202 pKa = 4.18
Molecular weight: 125.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q2GAX6|Q2GAX6_NOVAD GMP synthase [glutamine-hydrolyzing] OS=Novosphingobium aromaticivorans (strain ATCC 700278 / DSM 12444 / CCUG 56034 / CIP 105152 / NBRC 16084 / F199) OX=279238 GN=guaA PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.54GFRR19 pKa = 11.84ARR21 pKa = 11.84TATVGGRR28 pKa = 11.84KK29 pKa = 9.07VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.54GFRR19 pKa = 11.84ARR21 pKa = 11.84TATVGGRR28 pKa = 11.84KK29 pKa = 9.07VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1285425 |
41 |
3069 |
328.2 |
35.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.189 ± 0.047 | 0.877 ± 0.013 |
5.824 ± 0.027 | 5.649 ± 0.032 |
3.611 ± 0.024 | 9.085 ± 0.052 |
2.014 ± 0.02 | 4.683 ± 0.022 |
3.035 ± 0.033 | 9.727 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.486 ± 0.02 | 2.613 ± 0.023 |
5.324 ± 0.028 | 3.053 ± 0.02 |
7.303 ± 0.041 | 5.272 ± 0.033 |
5.181 ± 0.029 | 7.399 ± 0.029 |
1.484 ± 0.018 | 2.191 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
