
Emiliania huxleyi virus 99B1
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Algavirales; Phycodnaviridae; Coccolithovirus; unclassified Coccolithovirus
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
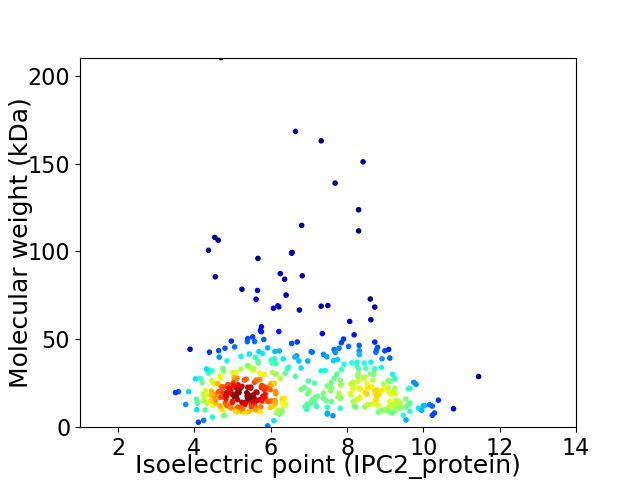
Virtual 2D-PAGE plot for 444 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
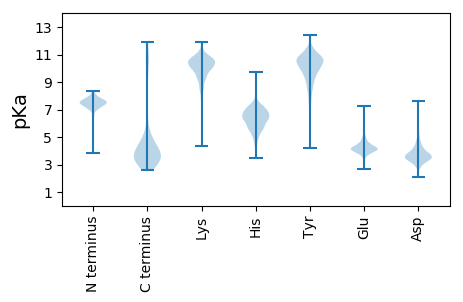
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D2TFC0|D2TFC0_9PHYC Putative C2H2-type Zinc finger protein OS=Emiliania huxleyi virus 99B1 OX=187397 GN=EhV184A PE=4 SV=1
MM1 pKa = 7.46LSKK4 pKa = 10.26FIHH7 pKa = 5.8MFCIGVSIAGYY18 pKa = 9.44PSGYY22 pKa = 9.55YY23 pKa = 10.19NRR25 pKa = 11.84LTPNAPNVGTFHH37 pKa = 6.49GTCTCPDD44 pKa = 3.17GSVYY48 pKa = 10.43IVGDD52 pKa = 3.51NVNQCGSLACYY63 pKa = 10.07GGVSGTCHH71 pKa = 6.81KK72 pKa = 11.22SNVAMDD78 pKa = 5.02HH79 pKa = 5.91IQGLMVEE86 pKa = 4.85CNVVAPSPPPSPPPSSPPSPSPPPSPLAPGHH117 pKa = 7.04DD118 pKa = 4.18YY119 pKa = 11.87VEE121 pKa = 4.52MFTSNISTSFIIAEE135 pKa = 4.69PDD137 pKa = 3.81LIDD140 pKa = 4.53PGSGSEE146 pKa = 3.85IGSGEE151 pKa = 3.88GRR153 pKa = 11.84RR154 pKa = 11.84LLEE157 pKa = 3.88TAAISPLVFEE167 pKa = 4.56YY168 pKa = 11.37AMYY171 pKa = 8.81TAFRR175 pKa = 11.84IMYY178 pKa = 7.24NVPRR182 pKa = 11.84FHH184 pKa = 7.65PDD186 pKa = 2.65CGLIEE191 pKa = 4.8SISNKK196 pKa = 10.17CIDD199 pKa = 3.25IMYY202 pKa = 10.67DD203 pKa = 3.27EE204 pKa = 5.27DD205 pKa = 4.07TTNSSVKK212 pKa = 10.37VCISPNMNSGDD223 pKa = 3.71TSITDD228 pKa = 3.66ALSTEE233 pKa = 4.41SGADD237 pKa = 3.66PIEE240 pKa = 4.97AIVDD244 pKa = 3.68IFNKK248 pKa = 10.13IIIGEE253 pKa = 4.23DD254 pKa = 3.02VGSSFFEE261 pKa = 4.22VSTIDD266 pKa = 3.98VPADD270 pKa = 3.81LYY272 pKa = 11.54AAFIATDD279 pKa = 3.8DD280 pKa = 4.13NNTLIYY286 pKa = 10.61NISTFTNRR294 pKa = 11.84YY295 pKa = 5.38NTEE298 pKa = 4.24PYY300 pKa = 10.81DD301 pKa = 5.01LDD303 pKa = 5.75DD304 pKa = 5.53PDD306 pKa = 3.68VTITFINPTSTQIVNKK322 pKa = 9.7PADD325 pKa = 4.79DD326 pKa = 6.11DD327 pKa = 6.37DD328 pKa = 7.46DD329 pKa = 7.38DD330 pKa = 7.43DD331 pKa = 7.58DD332 pKa = 7.0DD333 pKa = 4.9GLSDD337 pKa = 4.17GDD339 pKa = 3.68VAGIIIAAIIGLMFILIIAFVMWPKK364 pKa = 10.58NSRR367 pKa = 11.84VCVRR371 pKa = 11.84RR372 pKa = 11.84DD373 pKa = 3.32PEE375 pKa = 4.53VISTTPTPGLEE386 pKa = 4.28TVSPPTEE393 pKa = 3.97RR394 pKa = 11.84EE395 pKa = 3.82ITVATQPSAEE405 pKa = 4.16VTEE408 pKa = 4.36ITMTEE413 pKa = 3.99PP414 pKa = 3.64
MM1 pKa = 7.46LSKK4 pKa = 10.26FIHH7 pKa = 5.8MFCIGVSIAGYY18 pKa = 9.44PSGYY22 pKa = 9.55YY23 pKa = 10.19NRR25 pKa = 11.84LTPNAPNVGTFHH37 pKa = 6.49GTCTCPDD44 pKa = 3.17GSVYY48 pKa = 10.43IVGDD52 pKa = 3.51NVNQCGSLACYY63 pKa = 10.07GGVSGTCHH71 pKa = 6.81KK72 pKa = 11.22SNVAMDD78 pKa = 5.02HH79 pKa = 5.91IQGLMVEE86 pKa = 4.85CNVVAPSPPPSPPPSSPPSPSPPPSPLAPGHH117 pKa = 7.04DD118 pKa = 4.18YY119 pKa = 11.87VEE121 pKa = 4.52MFTSNISTSFIIAEE135 pKa = 4.69PDD137 pKa = 3.81LIDD140 pKa = 4.53PGSGSEE146 pKa = 3.85IGSGEE151 pKa = 3.88GRR153 pKa = 11.84RR154 pKa = 11.84LLEE157 pKa = 3.88TAAISPLVFEE167 pKa = 4.56YY168 pKa = 11.37AMYY171 pKa = 8.81TAFRR175 pKa = 11.84IMYY178 pKa = 7.24NVPRR182 pKa = 11.84FHH184 pKa = 7.65PDD186 pKa = 2.65CGLIEE191 pKa = 4.8SISNKK196 pKa = 10.17CIDD199 pKa = 3.25IMYY202 pKa = 10.67DD203 pKa = 3.27EE204 pKa = 5.27DD205 pKa = 4.07TTNSSVKK212 pKa = 10.37VCISPNMNSGDD223 pKa = 3.71TSITDD228 pKa = 3.66ALSTEE233 pKa = 4.41SGADD237 pKa = 3.66PIEE240 pKa = 4.97AIVDD244 pKa = 3.68IFNKK248 pKa = 10.13IIIGEE253 pKa = 4.23DD254 pKa = 3.02VGSSFFEE261 pKa = 4.22VSTIDD266 pKa = 3.98VPADD270 pKa = 3.81LYY272 pKa = 11.54AAFIATDD279 pKa = 3.8DD280 pKa = 4.13NNTLIYY286 pKa = 10.61NISTFTNRR294 pKa = 11.84YY295 pKa = 5.38NTEE298 pKa = 4.24PYY300 pKa = 10.81DD301 pKa = 5.01LDD303 pKa = 5.75DD304 pKa = 5.53PDD306 pKa = 3.68VTITFINPTSTQIVNKK322 pKa = 9.7PADD325 pKa = 4.79DD326 pKa = 6.11DD327 pKa = 6.37DD328 pKa = 7.46DD329 pKa = 7.38DD330 pKa = 7.43DD331 pKa = 7.58DD332 pKa = 7.0DD333 pKa = 4.9GLSDD337 pKa = 4.17GDD339 pKa = 3.68VAGIIIAAIIGLMFILIIAFVMWPKK364 pKa = 10.58NSRR367 pKa = 11.84VCVRR371 pKa = 11.84RR372 pKa = 11.84DD373 pKa = 3.32PEE375 pKa = 4.53VISTTPTPGLEE386 pKa = 4.28TVSPPTEE393 pKa = 3.97RR394 pKa = 11.84EE395 pKa = 3.82ITVATQPSAEE405 pKa = 4.16VTEE408 pKa = 4.36ITMTEE413 pKa = 3.99PP414 pKa = 3.64
Molecular weight: 44.34 kDa
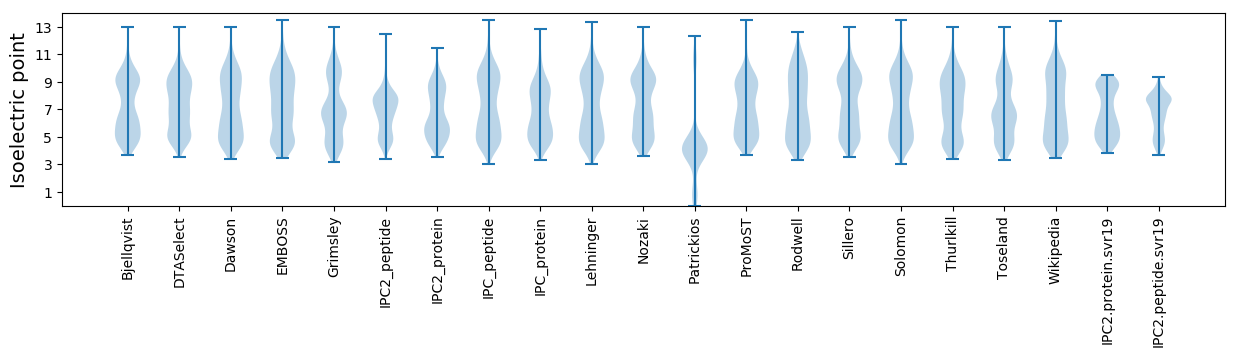
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D2TFC3|D2TFC3_9PHYC Putative membrane protein OS=Emiliania huxleyi virus 99B1 OX=187397 GN=EhV187 PE=4 SV=1
MM1 pKa = 7.25AVSRR5 pKa = 11.84RR6 pKa = 11.84SRR8 pKa = 11.84YY9 pKa = 7.36GTVRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84TVRR20 pKa = 11.84KK21 pKa = 8.12STASRR26 pKa = 11.84STKK29 pKa = 9.49VGRR32 pKa = 11.84VAKK35 pKa = 9.99LSNMSAGCRR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 10.33VKK48 pKa = 10.58SKK50 pKa = 10.47LGSRR54 pKa = 11.84KK55 pKa = 9.56LSRR58 pKa = 11.84ATRR61 pKa = 11.84ASIIRR66 pKa = 11.84SSVKK70 pKa = 8.95SWKK73 pKa = 10.17RR74 pKa = 11.84MSKK77 pKa = 7.59TRR79 pKa = 11.84KK80 pKa = 8.61AAYY83 pKa = 9.3SKK85 pKa = 10.35RR86 pKa = 11.84GRR88 pKa = 11.84VSTRR92 pKa = 11.84SRR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84STKK99 pKa = 8.68RR100 pKa = 11.84SRR102 pKa = 11.84RR103 pKa = 11.84TRR105 pKa = 11.84TKK107 pKa = 10.01RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84STKK113 pKa = 9.41RR114 pKa = 11.84SRR116 pKa = 11.84RR117 pKa = 11.84SSKK120 pKa = 9.88KK121 pKa = 9.43SRR123 pKa = 11.84RR124 pKa = 11.84STMKK128 pKa = 9.74RR129 pKa = 11.84PKK131 pKa = 10.03VSTMSKK137 pKa = 10.13SLKK140 pKa = 10.19KK141 pKa = 10.61YY142 pKa = 10.13IRR144 pKa = 11.84TKK146 pKa = 10.44SGKK149 pKa = 9.72KK150 pKa = 9.61NLSKK154 pKa = 11.04VSTKK158 pKa = 7.7MLRR161 pKa = 11.84KK162 pKa = 9.62LVALWRR168 pKa = 11.84RR169 pKa = 11.84MSAKK173 pKa = 10.19SKK175 pKa = 10.43AKK177 pKa = 9.76YY178 pKa = 9.51SVRR181 pKa = 11.84STRR184 pKa = 11.84KK185 pKa = 6.64STRR188 pKa = 11.84RR189 pKa = 11.84KK190 pKa = 6.68STRR193 pKa = 11.84RR194 pKa = 11.84KK195 pKa = 6.67STRR198 pKa = 11.84RR199 pKa = 11.84KK200 pKa = 7.3STRR203 pKa = 11.84KK204 pKa = 8.76RR205 pKa = 11.84RR206 pKa = 11.84RR207 pKa = 11.84SRR209 pKa = 11.84RR210 pKa = 11.84RR211 pKa = 11.84SRR213 pKa = 11.84PRR215 pKa = 11.84TRR217 pKa = 11.84RR218 pKa = 11.84VRR220 pKa = 11.84RR221 pKa = 11.84TRR223 pKa = 11.84TRR225 pKa = 11.84RR226 pKa = 11.84TKK228 pKa = 8.45TRR230 pKa = 11.84RR231 pKa = 11.84TRR233 pKa = 11.84RR234 pKa = 11.84TVRR237 pKa = 11.84RR238 pKa = 11.84RR239 pKa = 3.12
MM1 pKa = 7.25AVSRR5 pKa = 11.84RR6 pKa = 11.84SRR8 pKa = 11.84YY9 pKa = 7.36GTVRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84TVRR20 pKa = 11.84KK21 pKa = 8.12STASRR26 pKa = 11.84STKK29 pKa = 9.49VGRR32 pKa = 11.84VAKK35 pKa = 9.99LSNMSAGCRR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 10.33VKK48 pKa = 10.58SKK50 pKa = 10.47LGSRR54 pKa = 11.84KK55 pKa = 9.56LSRR58 pKa = 11.84ATRR61 pKa = 11.84ASIIRR66 pKa = 11.84SSVKK70 pKa = 8.95SWKK73 pKa = 10.17RR74 pKa = 11.84MSKK77 pKa = 7.59TRR79 pKa = 11.84KK80 pKa = 8.61AAYY83 pKa = 9.3SKK85 pKa = 10.35RR86 pKa = 11.84GRR88 pKa = 11.84VSTRR92 pKa = 11.84SRR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84STKK99 pKa = 8.68RR100 pKa = 11.84SRR102 pKa = 11.84RR103 pKa = 11.84TRR105 pKa = 11.84TKK107 pKa = 10.01RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84STKK113 pKa = 9.41RR114 pKa = 11.84SRR116 pKa = 11.84RR117 pKa = 11.84SSKK120 pKa = 9.88KK121 pKa = 9.43SRR123 pKa = 11.84RR124 pKa = 11.84STMKK128 pKa = 9.74RR129 pKa = 11.84PKK131 pKa = 10.03VSTMSKK137 pKa = 10.13SLKK140 pKa = 10.19KK141 pKa = 10.61YY142 pKa = 10.13IRR144 pKa = 11.84TKK146 pKa = 10.44SGKK149 pKa = 9.72KK150 pKa = 9.61NLSKK154 pKa = 11.04VSTKK158 pKa = 7.7MLRR161 pKa = 11.84KK162 pKa = 9.62LVALWRR168 pKa = 11.84RR169 pKa = 11.84MSAKK173 pKa = 10.19SKK175 pKa = 10.43AKK177 pKa = 9.76YY178 pKa = 9.51SVRR181 pKa = 11.84STRR184 pKa = 11.84KK185 pKa = 6.64STRR188 pKa = 11.84RR189 pKa = 11.84KK190 pKa = 6.68STRR193 pKa = 11.84RR194 pKa = 11.84KK195 pKa = 6.67STRR198 pKa = 11.84RR199 pKa = 11.84KK200 pKa = 7.3STRR203 pKa = 11.84KK204 pKa = 8.76RR205 pKa = 11.84RR206 pKa = 11.84RR207 pKa = 11.84SRR209 pKa = 11.84RR210 pKa = 11.84RR211 pKa = 11.84SRR213 pKa = 11.84PRR215 pKa = 11.84TRR217 pKa = 11.84RR218 pKa = 11.84VRR220 pKa = 11.84RR221 pKa = 11.84TRR223 pKa = 11.84TRR225 pKa = 11.84RR226 pKa = 11.84TKK228 pKa = 8.45TRR230 pKa = 11.84RR231 pKa = 11.84TRR233 pKa = 11.84RR234 pKa = 11.84TVRR237 pKa = 11.84RR238 pKa = 11.84RR239 pKa = 3.12
Molecular weight: 28.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
110970 |
8 |
1994 |
249.9 |
28.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.421 ± 0.114 | 1.814 ± 0.071 |
5.857 ± 0.1 | 5.146 ± 0.117 |
3.897 ± 0.075 | 5.275 ± 0.121 |
2.633 ± 0.07 | 7.34 ± 0.132 |
5.839 ± 0.152 | 7.471 ± 0.109 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.122 ± 0.076 | 5.626 ± 0.09 |
5.474 ± 0.305 | 3.09 ± 0.062 |
4.808 ± 0.111 | 7.295 ± 0.127 |
7.017 ± 0.095 | 6.506 ± 0.104 |
1.075 ± 0.039 | 4.295 ± 0.089 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
