
Consotaella salsifontis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Aurantimonadaceae; Consotaella
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
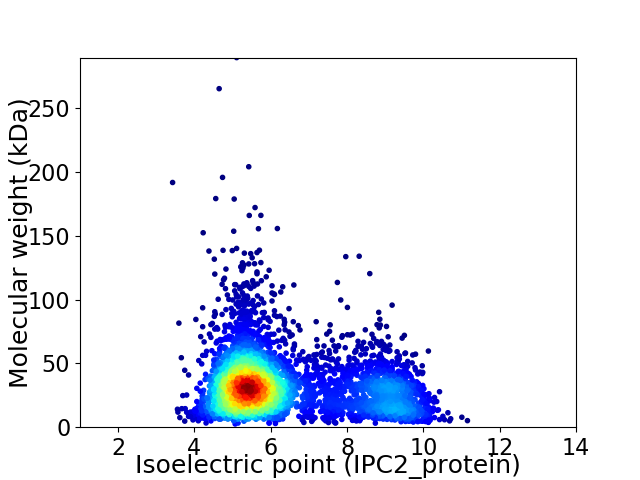
Virtual 2D-PAGE plot for 4266 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1T4P999|A0A1T4P999_9RHIZ Uncharacterized protein OS=Consotaella salsifontis OX=1365950 GN=SAMN05428963_103391 PE=4 SV=1
MM1 pKa = 7.47PVITGDD7 pKa = 3.25SGNNEE12 pKa = 4.21LVGTEE17 pKa = 3.97ADD19 pKa = 4.06DD20 pKa = 4.79VISGLDD26 pKa = 3.54GDD28 pKa = 5.51DD29 pKa = 4.53DD30 pKa = 5.21LSGLGGSDD38 pKa = 3.33TLDD41 pKa = 3.57GGDD44 pKa = 4.12GYY46 pKa = 11.5DD47 pKa = 3.48SIRR50 pKa = 11.84YY51 pKa = 8.48VGNSQGVVVTFTGQGSGTVEE71 pKa = 3.91EE72 pKa = 4.51NAIDD76 pKa = 3.72TDD78 pKa = 4.02SFTGIEE84 pKa = 4.32SVVGSQYY91 pKa = 11.6GDD93 pKa = 3.19VFTNSSSGYY102 pKa = 9.98ARR104 pKa = 11.84FRR106 pKa = 11.84GLDD109 pKa = 3.95GIDD112 pKa = 3.48TYY114 pKa = 11.59NGSEE118 pKa = 4.04TGFDD122 pKa = 3.52FLDD125 pKa = 4.13FLRR128 pKa = 11.84DD129 pKa = 3.47ANFGGTGGVTVDD141 pKa = 4.31LRR143 pKa = 11.84SGTAIDD149 pKa = 3.88GFGNLEE155 pKa = 4.01SLSYY159 pKa = 10.62IDD161 pKa = 3.81GARR164 pKa = 11.84GTEE167 pKa = 3.71QDD169 pKa = 4.35DD170 pKa = 4.16FFIGDD175 pKa = 3.93DD176 pKa = 3.84GDD178 pKa = 3.61NQFRR182 pKa = 11.84GEE184 pKa = 4.23GGNDD188 pKa = 3.3DD189 pKa = 3.62MRR191 pKa = 11.84ATGGSDD197 pKa = 3.33SFDD200 pKa = 3.31GGAGSDD206 pKa = 3.58SIQYY210 pKa = 10.79NDD212 pKa = 3.04ATQAIVAHH220 pKa = 5.53FTGDD224 pKa = 3.01LSGYY228 pKa = 7.81VLEE231 pKa = 5.12GSEE234 pKa = 4.24FTDD237 pKa = 3.07TFVDD241 pKa = 3.93VEE243 pKa = 4.61AVFGTAFGDD252 pKa = 3.76TFVNEE257 pKa = 4.26SSSFARR263 pKa = 11.84FRR265 pKa = 11.84GFDD268 pKa = 3.44GVDD271 pKa = 3.24SYY273 pKa = 11.55TGSASGFDD281 pKa = 3.78MIDD284 pKa = 3.93FYY286 pKa = 11.65RR287 pKa = 11.84DD288 pKa = 2.8ARR290 pKa = 11.84YY291 pKa = 10.24GGTSGVTVDD300 pKa = 4.43LAAGTGTDD308 pKa = 2.98GFGNVEE314 pKa = 4.28SFSNIDD320 pKa = 3.38GVRR323 pKa = 11.84GTDD326 pKa = 3.75FNDD329 pKa = 3.09VLTGSAGANTLDD341 pKa = 3.88GAGGADD347 pKa = 4.57LMRR350 pKa = 11.84GLAGDD355 pKa = 4.04DD356 pKa = 3.54VLVGSGADD364 pKa = 3.94GDD366 pKa = 4.21TADD369 pKa = 3.71YY370 pKa = 11.12SLDD373 pKa = 3.47VLYY376 pKa = 10.76GATHH380 pKa = 7.52GIRR383 pKa = 11.84VNQLGNGNQGGLPPDD398 pKa = 3.71TVIDD402 pKa = 3.78SFGYY406 pKa = 8.75TDD408 pKa = 4.07TIVDD412 pKa = 3.57VRR414 pKa = 11.84NIIATEE420 pKa = 3.86FDD422 pKa = 3.67DD423 pKa = 4.11EE424 pKa = 5.43VYY426 pKa = 10.96GGTHH430 pKa = 6.8GNTLVLGDD438 pKa = 3.94GDD440 pKa = 5.44DD441 pKa = 3.93YY442 pKa = 12.15AFGNAGNDD450 pKa = 3.55TLYY453 pKa = 11.21GGAGSDD459 pKa = 4.22RR460 pKa = 11.84MDD462 pKa = 3.94GGADD466 pKa = 3.29TDD468 pKa = 4.27TVVLGGLNGGGHH480 pKa = 7.62DD481 pKa = 4.06LLQFGGVVYY490 pKa = 10.78ALDD493 pKa = 4.14LRR495 pKa = 11.84DD496 pKa = 4.03HH497 pKa = 6.68SFDD500 pKa = 3.68TITNVEE506 pKa = 4.0QFQGVGQTIGLGAVEE521 pKa = 5.08GFDD524 pKa = 3.66PLVYY528 pKa = 10.17AASYY532 pKa = 10.98NDD534 pKa = 3.6LAQVFRR540 pKa = 11.84FDD542 pKa = 3.83YY543 pKa = 11.04NSALSHH549 pKa = 5.27YY550 pKa = 10.09VGTGYY555 pKa = 10.91FEE557 pKa = 4.46GRR559 pKa = 11.84EE560 pKa = 4.19ATFNAAQYY568 pKa = 9.23LANYY572 pKa = 9.6QDD574 pKa = 3.6LQAAFGTNYY583 pKa = 9.47EE584 pKa = 3.86AARR587 pKa = 11.84NHH589 pKa = 6.03FLASGVDD596 pKa = 3.54EE597 pKa = 4.87HH598 pKa = 7.52RR599 pKa = 11.84LAEE602 pKa = 4.74DD603 pKa = 3.57PLDD606 pKa = 4.29YY607 pKa = 10.48IASYY611 pKa = 11.17GDD613 pKa = 4.52LIQAFGGQSQSDD625 pKa = 3.77LVAAGLNHH633 pKa = 5.41YY634 pKa = 9.42RR635 pKa = 11.84AAGFDD640 pKa = 3.09EE641 pKa = 4.45GRR643 pKa = 11.84RR644 pKa = 11.84GGIDD648 pKa = 3.6FDD650 pKa = 3.74VGQYY654 pKa = 10.5LEE656 pKa = 4.89NYY658 pKa = 10.03GDD660 pKa = 3.75LQSAFGGDD668 pKa = 3.34GDD670 pKa = 4.61AATAHH675 pKa = 6.48YY676 pKa = 10.34INAGYY681 pKa = 10.45GEE683 pKa = 4.51HH684 pKa = 7.23RR685 pKa = 11.84LAEE688 pKa = 4.51DD689 pKa = 3.66PLDD692 pKa = 4.29YY693 pKa = 10.48IASYY697 pKa = 11.12GDD699 pKa = 3.96LIEE702 pKa = 5.37AFGSGNEE709 pKa = 4.0AQIIQAGLVHH719 pKa = 5.91YY720 pKa = 10.4QSGGWSEE727 pKa = 4.19GRR729 pKa = 11.84EE730 pKa = 3.67ADD732 pKa = 4.0FDD734 pKa = 4.21VDD736 pKa = 4.8SYY738 pKa = 11.36LANYY742 pKa = 9.69DD743 pKa = 3.64DD744 pKa = 5.23LRR746 pKa = 11.84AAFADD751 pKa = 4.24GAGGYY756 pKa = 9.85DD757 pKa = 3.44EE758 pKa = 5.38EE759 pKa = 5.3AALLHH764 pKa = 6.67FIHH767 pKa = 7.09AGYY770 pKa = 11.11AEE772 pKa = 4.34GRR774 pKa = 11.84TDD776 pKa = 5.22DD777 pKa = 5.36LMAAA781 pKa = 4.43
MM1 pKa = 7.47PVITGDD7 pKa = 3.25SGNNEE12 pKa = 4.21LVGTEE17 pKa = 3.97ADD19 pKa = 4.06DD20 pKa = 4.79VISGLDD26 pKa = 3.54GDD28 pKa = 5.51DD29 pKa = 4.53DD30 pKa = 5.21LSGLGGSDD38 pKa = 3.33TLDD41 pKa = 3.57GGDD44 pKa = 4.12GYY46 pKa = 11.5DD47 pKa = 3.48SIRR50 pKa = 11.84YY51 pKa = 8.48VGNSQGVVVTFTGQGSGTVEE71 pKa = 3.91EE72 pKa = 4.51NAIDD76 pKa = 3.72TDD78 pKa = 4.02SFTGIEE84 pKa = 4.32SVVGSQYY91 pKa = 11.6GDD93 pKa = 3.19VFTNSSSGYY102 pKa = 9.98ARR104 pKa = 11.84FRR106 pKa = 11.84GLDD109 pKa = 3.95GIDD112 pKa = 3.48TYY114 pKa = 11.59NGSEE118 pKa = 4.04TGFDD122 pKa = 3.52FLDD125 pKa = 4.13FLRR128 pKa = 11.84DD129 pKa = 3.47ANFGGTGGVTVDD141 pKa = 4.31LRR143 pKa = 11.84SGTAIDD149 pKa = 3.88GFGNLEE155 pKa = 4.01SLSYY159 pKa = 10.62IDD161 pKa = 3.81GARR164 pKa = 11.84GTEE167 pKa = 3.71QDD169 pKa = 4.35DD170 pKa = 4.16FFIGDD175 pKa = 3.93DD176 pKa = 3.84GDD178 pKa = 3.61NQFRR182 pKa = 11.84GEE184 pKa = 4.23GGNDD188 pKa = 3.3DD189 pKa = 3.62MRR191 pKa = 11.84ATGGSDD197 pKa = 3.33SFDD200 pKa = 3.31GGAGSDD206 pKa = 3.58SIQYY210 pKa = 10.79NDD212 pKa = 3.04ATQAIVAHH220 pKa = 5.53FTGDD224 pKa = 3.01LSGYY228 pKa = 7.81VLEE231 pKa = 5.12GSEE234 pKa = 4.24FTDD237 pKa = 3.07TFVDD241 pKa = 3.93VEE243 pKa = 4.61AVFGTAFGDD252 pKa = 3.76TFVNEE257 pKa = 4.26SSSFARR263 pKa = 11.84FRR265 pKa = 11.84GFDD268 pKa = 3.44GVDD271 pKa = 3.24SYY273 pKa = 11.55TGSASGFDD281 pKa = 3.78MIDD284 pKa = 3.93FYY286 pKa = 11.65RR287 pKa = 11.84DD288 pKa = 2.8ARR290 pKa = 11.84YY291 pKa = 10.24GGTSGVTVDD300 pKa = 4.43LAAGTGTDD308 pKa = 2.98GFGNVEE314 pKa = 4.28SFSNIDD320 pKa = 3.38GVRR323 pKa = 11.84GTDD326 pKa = 3.75FNDD329 pKa = 3.09VLTGSAGANTLDD341 pKa = 3.88GAGGADD347 pKa = 4.57LMRR350 pKa = 11.84GLAGDD355 pKa = 4.04DD356 pKa = 3.54VLVGSGADD364 pKa = 3.94GDD366 pKa = 4.21TADD369 pKa = 3.71YY370 pKa = 11.12SLDD373 pKa = 3.47VLYY376 pKa = 10.76GATHH380 pKa = 7.52GIRR383 pKa = 11.84VNQLGNGNQGGLPPDD398 pKa = 3.71TVIDD402 pKa = 3.78SFGYY406 pKa = 8.75TDD408 pKa = 4.07TIVDD412 pKa = 3.57VRR414 pKa = 11.84NIIATEE420 pKa = 3.86FDD422 pKa = 3.67DD423 pKa = 4.11EE424 pKa = 5.43VYY426 pKa = 10.96GGTHH430 pKa = 6.8GNTLVLGDD438 pKa = 3.94GDD440 pKa = 5.44DD441 pKa = 3.93YY442 pKa = 12.15AFGNAGNDD450 pKa = 3.55TLYY453 pKa = 11.21GGAGSDD459 pKa = 4.22RR460 pKa = 11.84MDD462 pKa = 3.94GGADD466 pKa = 3.29TDD468 pKa = 4.27TVVLGGLNGGGHH480 pKa = 7.62DD481 pKa = 4.06LLQFGGVVYY490 pKa = 10.78ALDD493 pKa = 4.14LRR495 pKa = 11.84DD496 pKa = 4.03HH497 pKa = 6.68SFDD500 pKa = 3.68TITNVEE506 pKa = 4.0QFQGVGQTIGLGAVEE521 pKa = 5.08GFDD524 pKa = 3.66PLVYY528 pKa = 10.17AASYY532 pKa = 10.98NDD534 pKa = 3.6LAQVFRR540 pKa = 11.84FDD542 pKa = 3.83YY543 pKa = 11.04NSALSHH549 pKa = 5.27YY550 pKa = 10.09VGTGYY555 pKa = 10.91FEE557 pKa = 4.46GRR559 pKa = 11.84EE560 pKa = 4.19ATFNAAQYY568 pKa = 9.23LANYY572 pKa = 9.6QDD574 pKa = 3.6LQAAFGTNYY583 pKa = 9.47EE584 pKa = 3.86AARR587 pKa = 11.84NHH589 pKa = 6.03FLASGVDD596 pKa = 3.54EE597 pKa = 4.87HH598 pKa = 7.52RR599 pKa = 11.84LAEE602 pKa = 4.74DD603 pKa = 3.57PLDD606 pKa = 4.29YY607 pKa = 10.48IASYY611 pKa = 11.17GDD613 pKa = 4.52LIQAFGGQSQSDD625 pKa = 3.77LVAAGLNHH633 pKa = 5.41YY634 pKa = 9.42RR635 pKa = 11.84AAGFDD640 pKa = 3.09EE641 pKa = 4.45GRR643 pKa = 11.84RR644 pKa = 11.84GGIDD648 pKa = 3.6FDD650 pKa = 3.74VGQYY654 pKa = 10.5LEE656 pKa = 4.89NYY658 pKa = 10.03GDD660 pKa = 3.75LQSAFGGDD668 pKa = 3.34GDD670 pKa = 4.61AATAHH675 pKa = 6.48YY676 pKa = 10.34INAGYY681 pKa = 10.45GEE683 pKa = 4.51HH684 pKa = 7.23RR685 pKa = 11.84LAEE688 pKa = 4.51DD689 pKa = 3.66PLDD692 pKa = 4.29YY693 pKa = 10.48IASYY697 pKa = 11.12GDD699 pKa = 3.96LIEE702 pKa = 5.37AFGSGNEE709 pKa = 4.0AQIIQAGLVHH719 pKa = 5.91YY720 pKa = 10.4QSGGWSEE727 pKa = 4.19GRR729 pKa = 11.84EE730 pKa = 3.67ADD732 pKa = 4.0FDD734 pKa = 4.21VDD736 pKa = 4.8SYY738 pKa = 11.36LANYY742 pKa = 9.69DD743 pKa = 3.64DD744 pKa = 5.23LRR746 pKa = 11.84AAFADD751 pKa = 4.24GAGGYY756 pKa = 9.85DD757 pKa = 3.44EE758 pKa = 5.38EE759 pKa = 5.3AALLHH764 pKa = 6.67FIHH767 pKa = 7.09AGYY770 pKa = 11.11AEE772 pKa = 4.34GRR774 pKa = 11.84TDD776 pKa = 5.22DD777 pKa = 5.36LMAAA781 pKa = 4.43
Molecular weight: 81.56 kDa
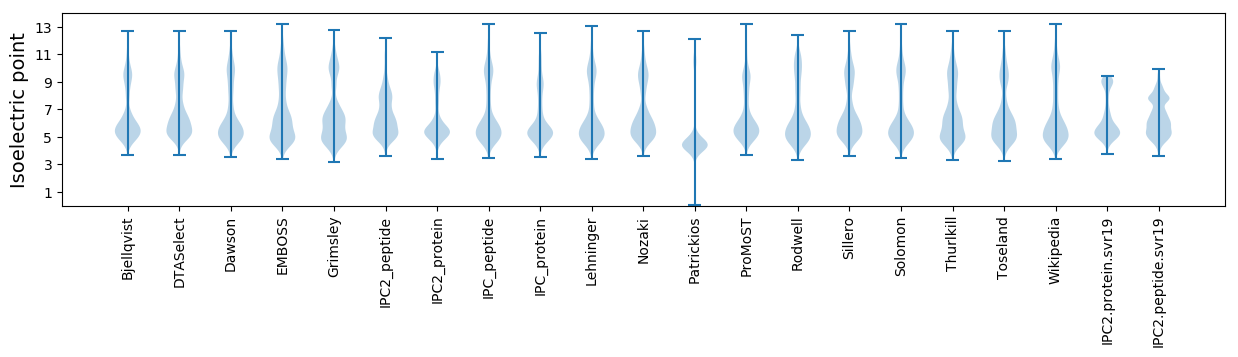
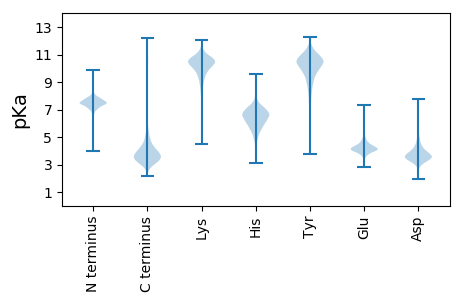
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1T4Q481|A0A1T4Q481_9RHIZ Methyltransferase domain-containing protein OS=Consotaella salsifontis OX=1365950 GN=SAMN05428963_104340 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATTGGRR28 pKa = 11.84KK29 pKa = 9.29VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATTGGRR28 pKa = 11.84KK29 pKa = 9.29VIAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1357347 |
24 |
2696 |
318.2 |
34.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.711 ± 0.049 | 0.826 ± 0.012 |
5.699 ± 0.033 | 6.199 ± 0.036 |
3.771 ± 0.024 | 8.504 ± 0.035 |
1.959 ± 0.019 | 5.2 ± 0.027 |
3.208 ± 0.028 | 10.083 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.464 ± 0.018 | 2.382 ± 0.021 |
5.098 ± 0.03 | 2.894 ± 0.024 |
7.325 ± 0.047 | 5.736 ± 0.027 |
5.122 ± 0.026 | 7.51 ± 0.031 |
1.227 ± 0.016 | 2.081 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
