
Klebsiella phage 48ST307
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
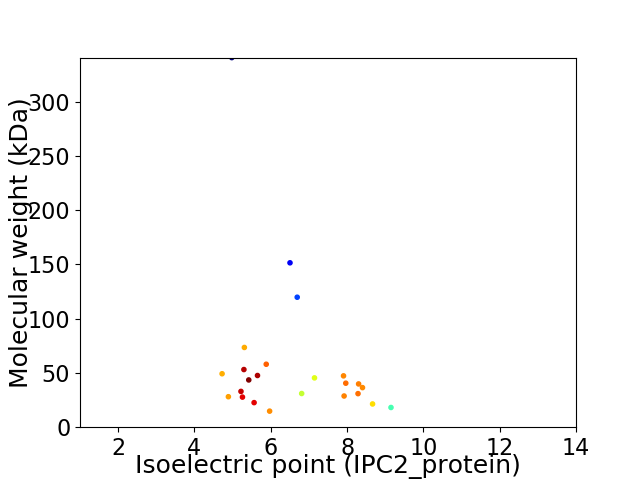
Virtual 2D-PAGE plot for 24 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z2EKP6|A0A2Z2EKP6_9CAUD DUF1983 domain-containing protein OS=Klebsiella phage 48ST307 OX=1955242 PE=4 SV=1
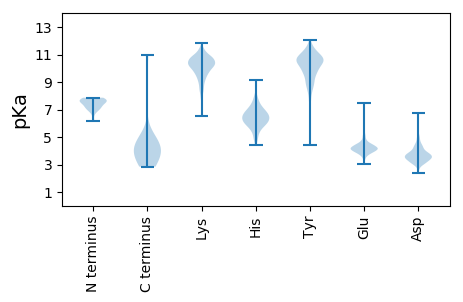
MM1 pKa = 7.23FCADD5 pKa = 4.25GEE7 pKa = 4.42NSAEE11 pKa = 4.41VYY13 pKa = 10.7CGATTMAQAKK23 pKa = 9.81KK24 pKa = 10.13VFTPARR30 pKa = 11.84QMASRR35 pKa = 11.84LPALRR40 pKa = 11.84SRR42 pKa = 11.84FDD44 pKa = 3.08ISVWTDD50 pKa = 2.52SLTRR54 pKa = 11.84PDD56 pKa = 3.85GSVFAPMAGKK66 pKa = 9.34PGDD69 pKa = 4.0GDD71 pKa = 4.02SPHH74 pKa = 6.8CAIIDD79 pKa = 3.92EE80 pKa = 4.45YY81 pKa = 11.24HH82 pKa = 6.38EE83 pKa = 5.19HH84 pKa = 6.91DD85 pKa = 3.78TDD87 pKa = 3.67HH88 pKa = 6.81MYY90 pKa = 11.12EE91 pKa = 3.95AMTMGMGARR100 pKa = 11.84SQPLTLIITTAGSSLEE116 pKa = 4.17SPCYY120 pKa = 10.68DD121 pKa = 3.04KK122 pKa = 11.39DD123 pKa = 4.18KK124 pKa = 10.73EE125 pKa = 4.36VKK127 pKa = 9.94EE128 pKa = 4.17VIEE131 pKa = 4.94GITRR135 pKa = 11.84NDD137 pKa = 3.29RR138 pKa = 11.84LFGMIYY144 pKa = 10.26EE145 pKa = 5.0LDD147 pKa = 4.53AGDD150 pKa = 5.23DD151 pKa = 3.4WTDD154 pKa = 3.32PKK156 pKa = 11.43NLIKK160 pKa = 10.8ANPNLDD166 pKa = 3.39VSVKK170 pKa = 10.55YY171 pKa = 10.59SDD173 pKa = 4.15LVEE176 pKa = 4.2LLEE179 pKa = 4.38VAKK182 pKa = 10.21QVPRR186 pKa = 11.84KK187 pKa = 10.19VNAFKK192 pKa = 10.48TKK194 pKa = 10.31RR195 pKa = 11.84LNIWVSGKK203 pKa = 9.62SAFYY207 pKa = 11.32NMEE210 pKa = 3.53QWKK213 pKa = 9.72AAEE216 pKa = 4.52DD217 pKa = 4.18PNLEE221 pKa = 4.3LADD224 pKa = 4.47FANDD228 pKa = 3.19SCNIGLDD235 pKa = 3.72LAKK238 pKa = 10.67KK239 pKa = 10.48LDD241 pKa = 3.71MNAGIRR247 pKa = 11.84LFTRR251 pKa = 11.84EE252 pKa = 3.9IEE254 pKa = 4.54GKK256 pKa = 9.13RR257 pKa = 11.84HH258 pKa = 5.31YY259 pKa = 11.03YY260 pKa = 9.68CIKK263 pKa = 9.43PKK265 pKa = 10.34FWVPEE270 pKa = 4.11DD271 pKa = 4.29TIHH274 pKa = 5.58TTDD277 pKa = 3.56PKK279 pKa = 10.94LLKK282 pKa = 9.73TADD285 pKa = 3.97RR286 pKa = 11.84YY287 pKa = 10.26QKK289 pKa = 9.95FYY291 pKa = 11.35EE292 pKa = 4.15MGVLEE297 pKa = 4.53ATDD300 pKa = 3.79GAEE303 pKa = 3.6ADD305 pKa = 3.69YY306 pKa = 11.36RR307 pKa = 11.84EE308 pKa = 4.26ILASIIDD315 pKa = 3.84MQDD318 pKa = 2.67EE319 pKa = 4.12NRR321 pKa = 11.84IDD323 pKa = 4.64EE324 pKa = 4.39IDD326 pKa = 3.76IDD328 pKa = 4.19PAGATALRR336 pKa = 11.84HH337 pKa = 4.74QLEE340 pKa = 4.81DD341 pKa = 3.17NGFTVVDD348 pKa = 3.54IRR350 pKa = 11.84QDD352 pKa = 3.6YY353 pKa = 9.7TNMSPAMKK361 pKa = 9.91EE362 pKa = 3.83LEE364 pKa = 4.0AALAGGRR371 pKa = 11.84FHH373 pKa = 7.84HH374 pKa = 7.44DD375 pKa = 3.32GNPILTWCISNVIGKK390 pKa = 7.4FIPGSDD396 pKa = 3.41DD397 pKa = 3.2LVRR400 pKa = 11.84PTKK403 pKa = 10.66GDD405 pKa = 3.49NQSKK409 pKa = 10.31IDD411 pKa = 3.58GATALFNAMTRR422 pKa = 11.84AMLHH426 pKa = 6.02EE427 pKa = 4.75SSGGTSVYY435 pKa = 10.66DD436 pKa = 4.16EE437 pKa = 4.82EE438 pKa = 7.47DD439 pKa = 3.34IACC442 pKa = 4.51
MM1 pKa = 7.23FCADD5 pKa = 4.25GEE7 pKa = 4.42NSAEE11 pKa = 4.41VYY13 pKa = 10.7CGATTMAQAKK23 pKa = 9.81KK24 pKa = 10.13VFTPARR30 pKa = 11.84QMASRR35 pKa = 11.84LPALRR40 pKa = 11.84SRR42 pKa = 11.84FDD44 pKa = 3.08ISVWTDD50 pKa = 2.52SLTRR54 pKa = 11.84PDD56 pKa = 3.85GSVFAPMAGKK66 pKa = 9.34PGDD69 pKa = 4.0GDD71 pKa = 4.02SPHH74 pKa = 6.8CAIIDD79 pKa = 3.92EE80 pKa = 4.45YY81 pKa = 11.24HH82 pKa = 6.38EE83 pKa = 5.19HH84 pKa = 6.91DD85 pKa = 3.78TDD87 pKa = 3.67HH88 pKa = 6.81MYY90 pKa = 11.12EE91 pKa = 3.95AMTMGMGARR100 pKa = 11.84SQPLTLIITTAGSSLEE116 pKa = 4.17SPCYY120 pKa = 10.68DD121 pKa = 3.04KK122 pKa = 11.39DD123 pKa = 4.18KK124 pKa = 10.73EE125 pKa = 4.36VKK127 pKa = 9.94EE128 pKa = 4.17VIEE131 pKa = 4.94GITRR135 pKa = 11.84NDD137 pKa = 3.29RR138 pKa = 11.84LFGMIYY144 pKa = 10.26EE145 pKa = 5.0LDD147 pKa = 4.53AGDD150 pKa = 5.23DD151 pKa = 3.4WTDD154 pKa = 3.32PKK156 pKa = 11.43NLIKK160 pKa = 10.8ANPNLDD166 pKa = 3.39VSVKK170 pKa = 10.55YY171 pKa = 10.59SDD173 pKa = 4.15LVEE176 pKa = 4.2LLEE179 pKa = 4.38VAKK182 pKa = 10.21QVPRR186 pKa = 11.84KK187 pKa = 10.19VNAFKK192 pKa = 10.48TKK194 pKa = 10.31RR195 pKa = 11.84LNIWVSGKK203 pKa = 9.62SAFYY207 pKa = 11.32NMEE210 pKa = 3.53QWKK213 pKa = 9.72AAEE216 pKa = 4.52DD217 pKa = 4.18PNLEE221 pKa = 4.3LADD224 pKa = 4.47FANDD228 pKa = 3.19SCNIGLDD235 pKa = 3.72LAKK238 pKa = 10.67KK239 pKa = 10.48LDD241 pKa = 3.71MNAGIRR247 pKa = 11.84LFTRR251 pKa = 11.84EE252 pKa = 3.9IEE254 pKa = 4.54GKK256 pKa = 9.13RR257 pKa = 11.84HH258 pKa = 5.31YY259 pKa = 11.03YY260 pKa = 9.68CIKK263 pKa = 9.43PKK265 pKa = 10.34FWVPEE270 pKa = 4.11DD271 pKa = 4.29TIHH274 pKa = 5.58TTDD277 pKa = 3.56PKK279 pKa = 10.94LLKK282 pKa = 9.73TADD285 pKa = 3.97RR286 pKa = 11.84YY287 pKa = 10.26QKK289 pKa = 9.95FYY291 pKa = 11.35EE292 pKa = 4.15MGVLEE297 pKa = 4.53ATDD300 pKa = 3.79GAEE303 pKa = 3.6ADD305 pKa = 3.69YY306 pKa = 11.36RR307 pKa = 11.84EE308 pKa = 4.26ILASIIDD315 pKa = 3.84MQDD318 pKa = 2.67EE319 pKa = 4.12NRR321 pKa = 11.84IDD323 pKa = 4.64EE324 pKa = 4.39IDD326 pKa = 3.76IDD328 pKa = 4.19PAGATALRR336 pKa = 11.84HH337 pKa = 4.74QLEE340 pKa = 4.81DD341 pKa = 3.17NGFTVVDD348 pKa = 3.54IRR350 pKa = 11.84QDD352 pKa = 3.6YY353 pKa = 9.7TNMSPAMKK361 pKa = 9.91EE362 pKa = 3.83LEE364 pKa = 4.0AALAGGRR371 pKa = 11.84FHH373 pKa = 7.84HH374 pKa = 7.44DD375 pKa = 3.32GNPILTWCISNVIGKK390 pKa = 7.4FIPGSDD396 pKa = 3.41DD397 pKa = 3.2LVRR400 pKa = 11.84PTKK403 pKa = 10.66GDD405 pKa = 3.49NQSKK409 pKa = 10.31IDD411 pKa = 3.58GATALFNAMTRR422 pKa = 11.84AMLHH426 pKa = 6.02EE427 pKa = 4.75SSGGTSVYY435 pKa = 10.66DD436 pKa = 4.16EE437 pKa = 4.82EE438 pKa = 7.47DD439 pKa = 3.34IACC442 pKa = 4.51
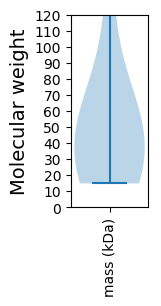
Molecular weight: 49.24 kDa
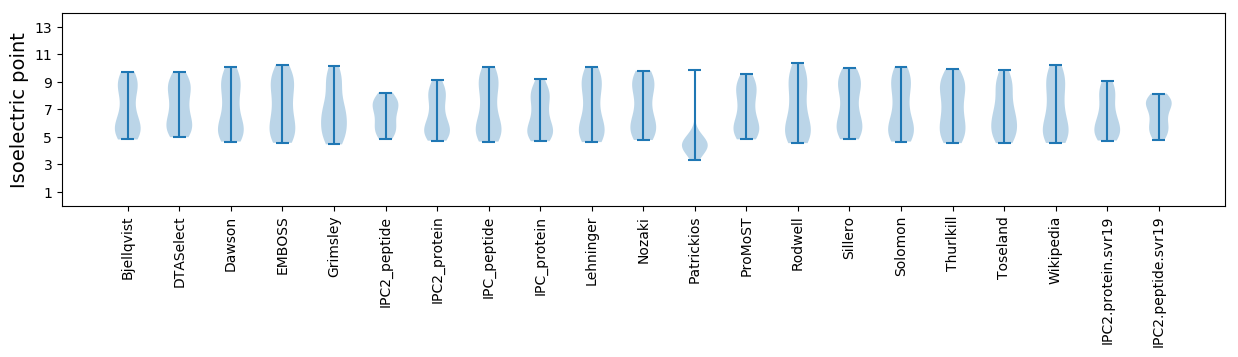
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z2EN75|A0A2Z2EN75_9CAUD Uncharacterized protein OS=Klebsiella phage 48ST307 OX=1955242 PE=4 SV=1
MM1 pKa = 7.86RR2 pKa = 11.84YY3 pKa = 9.47SRR5 pKa = 11.84SEE7 pKa = 3.62ARR9 pKa = 11.84GVIMNSLKK17 pKa = 10.67RR18 pKa = 11.84MAKK21 pKa = 9.82AWLLMNGAFVLLVMATQPAMANEE44 pKa = 4.19SQSLGLDD51 pKa = 3.19LDD53 pKa = 5.0SILSALPAGWASGVTAVFIVLYY75 pKa = 10.65AVAQLRR81 pKa = 11.84AVLPPSVTKK90 pKa = 10.82RR91 pKa = 11.84IPTVVMKK98 pKa = 10.78ILDD101 pKa = 3.89LVAANYY107 pKa = 9.88AHH109 pKa = 7.12ARR111 pKa = 11.84NADD114 pKa = 4.16AISKK118 pKa = 7.93VARR121 pKa = 11.84DD122 pKa = 3.38AGKK125 pKa = 10.49AKK127 pKa = 10.44GPSDD131 pKa = 2.95VDD133 pKa = 3.23YY134 pKa = 11.1RR135 pKa = 11.84VMVEE139 pKa = 3.78TAKK142 pKa = 11.17NNGEE146 pKa = 3.95LRR148 pKa = 11.84GSRR151 pKa = 11.84IEE153 pKa = 4.21SAGDD157 pKa = 3.41YY158 pKa = 10.78PGDD161 pKa = 4.03DD162 pKa = 3.63RR163 pKa = 11.84PGSKK167 pKa = 10.13SPQQ170 pKa = 3.11
MM1 pKa = 7.86RR2 pKa = 11.84YY3 pKa = 9.47SRR5 pKa = 11.84SEE7 pKa = 3.62ARR9 pKa = 11.84GVIMNSLKK17 pKa = 10.67RR18 pKa = 11.84MAKK21 pKa = 9.82AWLLMNGAFVLLVMATQPAMANEE44 pKa = 4.19SQSLGLDD51 pKa = 3.19LDD53 pKa = 5.0SILSALPAGWASGVTAVFIVLYY75 pKa = 10.65AVAQLRR81 pKa = 11.84AVLPPSVTKK90 pKa = 10.82RR91 pKa = 11.84IPTVVMKK98 pKa = 10.78ILDD101 pKa = 3.89LVAANYY107 pKa = 9.88AHH109 pKa = 7.12ARR111 pKa = 11.84NADD114 pKa = 4.16AISKK118 pKa = 7.93VARR121 pKa = 11.84DD122 pKa = 3.38AGKK125 pKa = 10.49AKK127 pKa = 10.44GPSDD131 pKa = 2.95VDD133 pKa = 3.23YY134 pKa = 11.1RR135 pKa = 11.84VMVEE139 pKa = 3.78TAKK142 pKa = 11.17NNGEE146 pKa = 3.95LRR148 pKa = 11.84GSRR151 pKa = 11.84IEE153 pKa = 4.21SAGDD157 pKa = 3.41YY158 pKa = 10.78PGDD161 pKa = 4.03DD162 pKa = 3.63RR163 pKa = 11.84PGSKK167 pKa = 10.13SPQQ170 pKa = 3.11
Molecular weight: 18.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12793 |
129 |
3193 |
533.0 |
58.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.177 ± 0.537 | 0.883 ± 0.19 |
5.925 ± 0.264 | 5.605 ± 0.518 |
3.088 ± 0.315 | 7.574 ± 0.408 |
1.423 ± 0.291 | 5.37 ± 0.295 |
5.143 ± 0.505 | 8.45 ± 0.434 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.415 ± 0.338 | 5.042 ± 0.455 |
3.572 ± 0.305 | 4.823 ± 0.366 |
5.495 ± 0.405 | 7.41 ± 0.695 |
6.98 ± 0.839 | 6.207 ± 0.367 |
1.438 ± 0.198 | 2.978 ± 0.181 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
