
Pichia membranifaciens
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Pichiaceae; Pichia
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
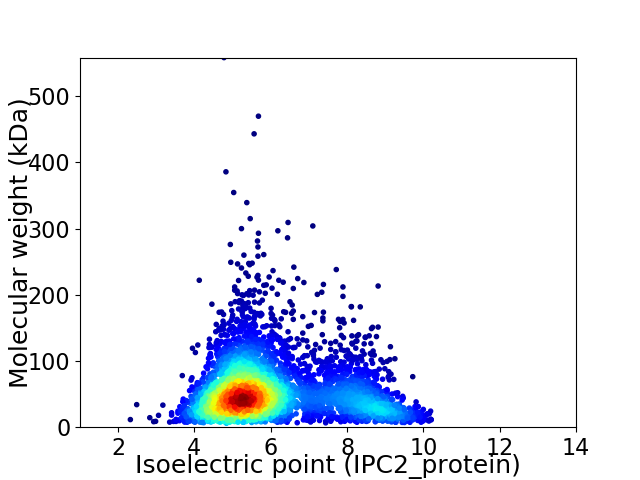
Virtual 2D-PAGE plot for 4160 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q2YCS2|A0A1Q2YCS2_9ASCO J domain-containing protein OS=Pichia membranifaciens OX=4926 GN=PMKS-000762 PE=4 SV=1
MM1 pKa = 7.14MMSSYY6 pKa = 10.34FQSTYY11 pKa = 10.07NSGRR15 pKa = 11.84IAQTYY20 pKa = 10.23LLASKK25 pKa = 10.46VKK27 pKa = 10.81AKK29 pKa = 8.93LTQEE33 pKa = 3.95AVRR36 pKa = 11.84NDD38 pKa = 3.29VDD40 pKa = 3.6LHH42 pKa = 7.25RR43 pKa = 11.84LVCQANLLDD52 pKa = 4.82NLIEE56 pKa = 4.25NLNHH60 pKa = 7.02HH61 pKa = 6.28EE62 pKa = 4.46SSYY65 pKa = 11.87NNNNSNTTGNTNICYY80 pKa = 10.24EE81 pKa = 4.55SINSNSYY88 pKa = 9.74PLLNNVSVQVNNLDD102 pKa = 3.61YY103 pKa = 11.34SVDD106 pKa = 3.7EE107 pKa = 5.54DD108 pKa = 5.5ADD110 pKa = 3.84SDD112 pKa = 5.02DD113 pKa = 4.63NDD115 pKa = 4.01EE116 pKa = 4.81EE117 pKa = 5.51DD118 pKa = 5.95DD119 pKa = 4.3EE120 pKa = 5.66VDD122 pKa = 3.56EE123 pKa = 4.6NDD125 pKa = 3.6IKK127 pKa = 10.8IEE129 pKa = 3.94RR130 pKa = 11.84APCSDD135 pKa = 3.3VYY137 pKa = 10.22YY138 pKa = 10.53TSDD141 pKa = 4.79DD142 pKa = 3.7SDD144 pKa = 3.98FDD146 pKa = 4.97SDD148 pKa = 4.91DD149 pKa = 5.19DD150 pKa = 4.68SDD152 pKa = 4.55SSFNSDD158 pKa = 2.94SFGGKK163 pKa = 9.74SSDD166 pKa = 3.94PNEE169 pKa = 4.3LCCIALQRR177 pKa = 11.84LNLHH181 pKa = 6.38GGVDD185 pKa = 4.02EE186 pKa = 6.84DD187 pKa = 5.48IDD189 pKa = 6.02DD190 pKa = 5.09DD191 pKa = 5.69DD192 pKa = 5.81SGNQEE197 pKa = 3.79NCSSVTATITLTDD210 pKa = 4.04SEE212 pKa = 4.96SDD214 pKa = 3.17SDD216 pKa = 4.51SNSEE220 pKa = 4.19TEE222 pKa = 4.71SDD224 pKa = 3.38TEE226 pKa = 4.23YY227 pKa = 11.08EE228 pKa = 3.95NDD230 pKa = 3.51SRR232 pKa = 11.84DD233 pKa = 3.6GLFSNNYY240 pKa = 7.46CALVRR245 pKa = 11.84MHH247 pKa = 6.32SQHH250 pKa = 6.45TSLNLNDD257 pKa = 3.78YY258 pKa = 9.46VASLEE263 pKa = 4.07ADD265 pKa = 3.48TSVEE269 pKa = 4.05EE270 pKa = 4.22TDD272 pKa = 3.31SCKK275 pKa = 10.7NIQHH279 pKa = 6.21TSSLDD284 pKa = 3.25EE285 pKa = 4.0MTRR288 pKa = 11.84EE289 pKa = 3.8EE290 pKa = 4.84DD291 pKa = 4.46LPSLSNCSSFSSMEE305 pKa = 4.55EE306 pKa = 3.2ISQPNQNIVRR316 pKa = 11.84LEE318 pKa = 4.09CLDD321 pKa = 3.88SIKK324 pKa = 10.23PQRR327 pKa = 11.84DD328 pKa = 2.99TDD330 pKa = 4.03SPSLYY335 pKa = 9.2LTQSQDD341 pKa = 2.98VFLLL345 pKa = 3.98
MM1 pKa = 7.14MMSSYY6 pKa = 10.34FQSTYY11 pKa = 10.07NSGRR15 pKa = 11.84IAQTYY20 pKa = 10.23LLASKK25 pKa = 10.46VKK27 pKa = 10.81AKK29 pKa = 8.93LTQEE33 pKa = 3.95AVRR36 pKa = 11.84NDD38 pKa = 3.29VDD40 pKa = 3.6LHH42 pKa = 7.25RR43 pKa = 11.84LVCQANLLDD52 pKa = 4.82NLIEE56 pKa = 4.25NLNHH60 pKa = 7.02HH61 pKa = 6.28EE62 pKa = 4.46SSYY65 pKa = 11.87NNNNSNTTGNTNICYY80 pKa = 10.24EE81 pKa = 4.55SINSNSYY88 pKa = 9.74PLLNNVSVQVNNLDD102 pKa = 3.61YY103 pKa = 11.34SVDD106 pKa = 3.7EE107 pKa = 5.54DD108 pKa = 5.5ADD110 pKa = 3.84SDD112 pKa = 5.02DD113 pKa = 4.63NDD115 pKa = 4.01EE116 pKa = 4.81EE117 pKa = 5.51DD118 pKa = 5.95DD119 pKa = 4.3EE120 pKa = 5.66VDD122 pKa = 3.56EE123 pKa = 4.6NDD125 pKa = 3.6IKK127 pKa = 10.8IEE129 pKa = 3.94RR130 pKa = 11.84APCSDD135 pKa = 3.3VYY137 pKa = 10.22YY138 pKa = 10.53TSDD141 pKa = 4.79DD142 pKa = 3.7SDD144 pKa = 3.98FDD146 pKa = 4.97SDD148 pKa = 4.91DD149 pKa = 5.19DD150 pKa = 4.68SDD152 pKa = 4.55SSFNSDD158 pKa = 2.94SFGGKK163 pKa = 9.74SSDD166 pKa = 3.94PNEE169 pKa = 4.3LCCIALQRR177 pKa = 11.84LNLHH181 pKa = 6.38GGVDD185 pKa = 4.02EE186 pKa = 6.84DD187 pKa = 5.48IDD189 pKa = 6.02DD190 pKa = 5.09DD191 pKa = 5.69DD192 pKa = 5.81SGNQEE197 pKa = 3.79NCSSVTATITLTDD210 pKa = 4.04SEE212 pKa = 4.96SDD214 pKa = 3.17SDD216 pKa = 4.51SNSEE220 pKa = 4.19TEE222 pKa = 4.71SDD224 pKa = 3.38TEE226 pKa = 4.23YY227 pKa = 11.08EE228 pKa = 3.95NDD230 pKa = 3.51SRR232 pKa = 11.84DD233 pKa = 3.6GLFSNNYY240 pKa = 7.46CALVRR245 pKa = 11.84MHH247 pKa = 6.32SQHH250 pKa = 6.45TSLNLNDD257 pKa = 3.78YY258 pKa = 9.46VASLEE263 pKa = 4.07ADD265 pKa = 3.48TSVEE269 pKa = 4.05EE270 pKa = 4.22TDD272 pKa = 3.31SCKK275 pKa = 10.7NIQHH279 pKa = 6.21TSSLDD284 pKa = 3.25EE285 pKa = 4.0MTRR288 pKa = 11.84EE289 pKa = 3.8EE290 pKa = 4.84DD291 pKa = 4.46LPSLSNCSSFSSMEE305 pKa = 4.55EE306 pKa = 3.2ISQPNQNIVRR316 pKa = 11.84LEE318 pKa = 4.09CLDD321 pKa = 3.88SIKK324 pKa = 10.23PQRR327 pKa = 11.84DD328 pKa = 2.99TDD330 pKa = 4.03SPSLYY335 pKa = 9.2LTQSQDD341 pKa = 2.98VFLLL345 pKa = 3.98
Molecular weight: 38.47 kDa
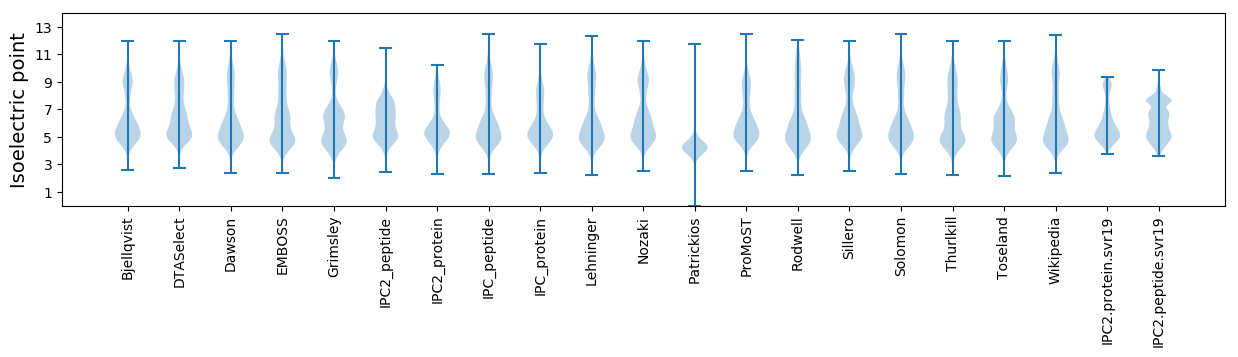
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q2YIB5|A0A1Q2YIB5_9ASCO DNA-directed RNA polymerases I II and III subunit RPABC1 OS=Pichia membranifaciens OX=4926 GN=PMKS-002769 PE=3 SV=1
MM1 pKa = 7.46ISARR5 pKa = 11.84SIKK8 pKa = 10.29AAAKK12 pKa = 9.64QALAVNARR20 pKa = 11.84IVPRR24 pKa = 11.84LVRR27 pKa = 11.84AYY29 pKa = 9.85HH30 pKa = 5.58QRR32 pKa = 11.84AQGGEE37 pKa = 3.91EE38 pKa = 4.12RR39 pKa = 11.84IAAAACSTRR48 pKa = 11.84ARR50 pKa = 11.84PLAARR55 pKa = 11.84TYY57 pKa = 10.85ASAKK61 pKa = 9.17AQPTEE66 pKa = 3.82VSSILEE72 pKa = 3.96SKK74 pKa = 10.2IRR76 pKa = 11.84GVSEE80 pKa = 3.66EE81 pKa = 4.35ANLDD85 pKa = 3.29EE86 pKa = 4.69TGRR89 pKa = 11.84VLAVGYY95 pKa = 10.47VLLSQQ100 pKa = 4.29
MM1 pKa = 7.46ISARR5 pKa = 11.84SIKK8 pKa = 10.29AAAKK12 pKa = 9.64QALAVNARR20 pKa = 11.84IVPRR24 pKa = 11.84LVRR27 pKa = 11.84AYY29 pKa = 9.85HH30 pKa = 5.58QRR32 pKa = 11.84AQGGEE37 pKa = 3.91EE38 pKa = 4.12RR39 pKa = 11.84IAAAACSTRR48 pKa = 11.84ARR50 pKa = 11.84PLAARR55 pKa = 11.84TYY57 pKa = 10.85ASAKK61 pKa = 9.17AQPTEE66 pKa = 3.82VSSILEE72 pKa = 3.96SKK74 pKa = 10.2IRR76 pKa = 11.84GVSEE80 pKa = 3.66EE81 pKa = 4.35ANLDD85 pKa = 3.29EE86 pKa = 4.69TGRR89 pKa = 11.84VLAVGYY95 pKa = 10.47VLLSQQ100 pKa = 4.29
Molecular weight: 10.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2159257 |
66 |
4922 |
519.1 |
58.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.003 ± 0.037 | 1.115 ± 0.012 |
6.429 ± 0.033 | 7.073 ± 0.039 |
4.213 ± 0.027 | 5.383 ± 0.033 |
1.993 ± 0.014 | 6.203 ± 0.033 |
7.442 ± 0.035 | 9.175 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.08 ± 0.013 | 6.068 ± 0.041 |
4.218 ± 0.031 | 3.803 ± 0.033 |
4.174 ± 0.025 | 9.076 ± 0.053 |
5.511 ± 0.029 | 5.817 ± 0.025 |
0.922 ± 0.011 | 3.3 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
