
Raccoon-associated polyomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Deltapolyomavirus; Procyon lotor polyomavirus 2
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
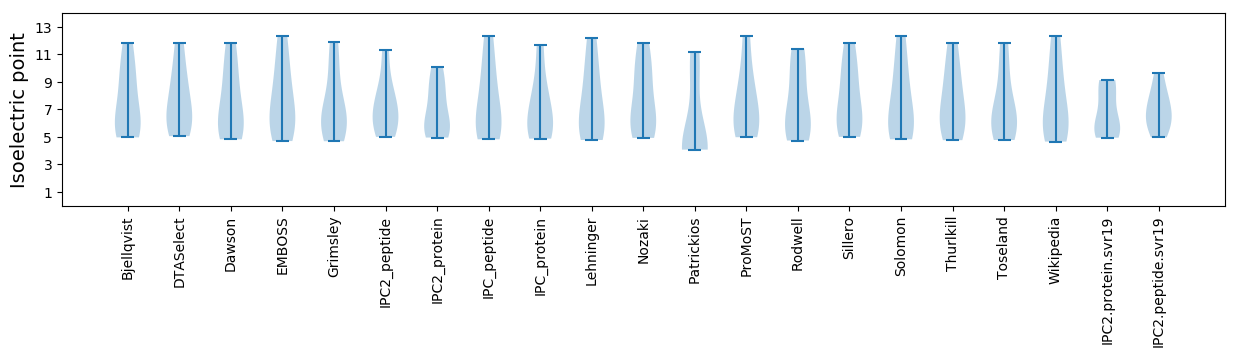
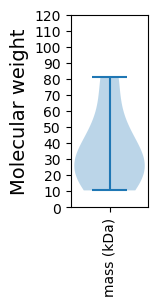
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1W5X9W6|A0A1W5X9W6_9POLY MT OS=Raccoon-associated polyomavirus 2 OX=1978920 PE=4 SV=1
MM1 pKa = 7.37GGVLSVLVDD10 pKa = 5.05LIQLLPEE17 pKa = 4.7LSASTGFGIEE27 pKa = 4.44AILAGEE33 pKa = 4.15AAASVEE39 pKa = 4.1AQVTSLMLVEE49 pKa = 4.74SLGPLDD55 pKa = 4.78ALASLGLSAEE65 pKa = 4.32SFSLLSAMPGFFQDD79 pKa = 3.73VVGLGVLFQTVSGASSLVAAGIQLRR104 pKa = 11.84HH105 pKa = 5.64EE106 pKa = 4.32VSVVNRR112 pKa = 11.84NMALAPWIPHH122 pKa = 6.69DD123 pKa = 4.9LYY125 pKa = 11.08DD126 pKa = 3.46IYY128 pKa = 11.13FPGVSTFSYY137 pKa = 9.25VVNVITDD144 pKa = 3.31WGISLFHH151 pKa = 6.61TIARR155 pKa = 11.84NVWDD159 pKa = 3.61EE160 pKa = 3.76LVRR163 pKa = 11.84EE164 pKa = 4.21GRR166 pKa = 11.84EE167 pKa = 3.74QIEE170 pKa = 4.15QVTRR174 pKa = 11.84DD175 pKa = 3.18VALRR179 pKa = 11.84GVQTFADD186 pKa = 3.68TMARR190 pKa = 11.84LIEE193 pKa = 4.03RR194 pKa = 11.84ARR196 pKa = 11.84WTITNSPIHH205 pKa = 5.68HH206 pKa = 7.09AYY208 pKa = 9.13WYY210 pKa = 10.99LEE212 pKa = 4.2DD213 pKa = 3.95YY214 pKa = 10.66YY215 pKa = 11.65KK216 pKa = 10.68RR217 pKa = 11.84LPQINPPQARR227 pKa = 11.84QLFRR231 pKa = 11.84RR232 pKa = 11.84IGQRR236 pKa = 11.84PPDD239 pKa = 3.61RR240 pKa = 11.84SNLEE244 pKa = 3.92EE245 pKa = 4.24LEE247 pKa = 4.19KK248 pKa = 10.74EE249 pKa = 4.09SRR251 pKa = 11.84EE252 pKa = 3.8EE253 pKa = 3.59SGQIVEE259 pKa = 4.76KK260 pKa = 10.97YY261 pKa = 8.77EE262 pKa = 4.03PPGGAFQRR270 pKa = 11.84VTPDD274 pKa = 2.31WMLPLILGLYY284 pKa = 10.53GDD286 pKa = 4.64ITPTWSTYY294 pKa = 10.0IEE296 pKa = 4.12QIEE299 pKa = 4.23AEE301 pKa = 3.95EE302 pKa = 4.83DD303 pKa = 3.81GPKK306 pKa = 10.06KK307 pKa = 10.09KK308 pKa = 9.98RR309 pKa = 11.84RR310 pKa = 11.84RR311 pKa = 11.84QQ312 pKa = 3.19
MM1 pKa = 7.37GGVLSVLVDD10 pKa = 5.05LIQLLPEE17 pKa = 4.7LSASTGFGIEE27 pKa = 4.44AILAGEE33 pKa = 4.15AAASVEE39 pKa = 4.1AQVTSLMLVEE49 pKa = 4.74SLGPLDD55 pKa = 4.78ALASLGLSAEE65 pKa = 4.32SFSLLSAMPGFFQDD79 pKa = 3.73VVGLGVLFQTVSGASSLVAAGIQLRR104 pKa = 11.84HH105 pKa = 5.64EE106 pKa = 4.32VSVVNRR112 pKa = 11.84NMALAPWIPHH122 pKa = 6.69DD123 pKa = 4.9LYY125 pKa = 11.08DD126 pKa = 3.46IYY128 pKa = 11.13FPGVSTFSYY137 pKa = 9.25VVNVITDD144 pKa = 3.31WGISLFHH151 pKa = 6.61TIARR155 pKa = 11.84NVWDD159 pKa = 3.61EE160 pKa = 3.76LVRR163 pKa = 11.84EE164 pKa = 4.21GRR166 pKa = 11.84EE167 pKa = 3.74QIEE170 pKa = 4.15QVTRR174 pKa = 11.84DD175 pKa = 3.18VALRR179 pKa = 11.84GVQTFADD186 pKa = 3.68TMARR190 pKa = 11.84LIEE193 pKa = 4.03RR194 pKa = 11.84ARR196 pKa = 11.84WTITNSPIHH205 pKa = 5.68HH206 pKa = 7.09AYY208 pKa = 9.13WYY210 pKa = 10.99LEE212 pKa = 4.2DD213 pKa = 3.95YY214 pKa = 10.66YY215 pKa = 11.65KK216 pKa = 10.68RR217 pKa = 11.84LPQINPPQARR227 pKa = 11.84QLFRR231 pKa = 11.84RR232 pKa = 11.84IGQRR236 pKa = 11.84PPDD239 pKa = 3.61RR240 pKa = 11.84SNLEE244 pKa = 3.92EE245 pKa = 4.24LEE247 pKa = 4.19KK248 pKa = 10.74EE249 pKa = 4.09SRR251 pKa = 11.84EE252 pKa = 3.8EE253 pKa = 3.59SGQIVEE259 pKa = 4.76KK260 pKa = 10.97YY261 pKa = 8.77EE262 pKa = 4.03PPGGAFQRR270 pKa = 11.84VTPDD274 pKa = 2.31WMLPLILGLYY284 pKa = 10.53GDD286 pKa = 4.64ITPTWSTYY294 pKa = 10.0IEE296 pKa = 4.12QIEE299 pKa = 4.23AEE301 pKa = 3.95EE302 pKa = 4.83DD303 pKa = 3.81GPKK306 pKa = 10.06KK307 pKa = 10.09KK308 pKa = 9.98RR309 pKa = 11.84RR310 pKa = 11.84RR311 pKa = 11.84QQ312 pKa = 3.19
Molecular weight: 34.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W5X9X0|A0A1W5X9X0_9POLY ALTO OS=Raccoon-associated polyomavirus 2 OX=1978920 PE=4 SV=1
MM1 pKa = 8.0RR2 pKa = 11.84NTSLHH7 pKa = 6.26PGVPEE12 pKa = 4.0APEE15 pKa = 4.17PLLLSLVLLLLPLPLLPHH33 pKa = 6.91PLLLQLILLLLLHH46 pKa = 6.91LGGLHH51 pKa = 6.75LCCIPPHH58 pKa = 6.39RR59 pKa = 11.84RR60 pKa = 11.84PPTTTRR66 pKa = 11.84EE67 pKa = 3.69RR68 pKa = 11.84SHH70 pKa = 6.5RR71 pKa = 11.84HH72 pKa = 4.38QLPSRR77 pKa = 11.84ASPHH81 pKa = 5.37PRR83 pKa = 11.84SRR85 pKa = 11.84RR86 pKa = 11.84KK87 pKa = 8.88WMHH90 pKa = 5.89HH91 pKa = 4.94
MM1 pKa = 8.0RR2 pKa = 11.84NTSLHH7 pKa = 6.26PGVPEE12 pKa = 4.0APEE15 pKa = 4.17PLLLSLVLLLLPLPLLPHH33 pKa = 6.91PLLLQLILLLLLHH46 pKa = 6.91LGGLHH51 pKa = 6.75LCCIPPHH58 pKa = 6.39RR59 pKa = 11.84RR60 pKa = 11.84PPTTTRR66 pKa = 11.84EE67 pKa = 3.69RR68 pKa = 11.84SHH70 pKa = 6.5RR71 pKa = 11.84HH72 pKa = 4.38QLPSRR77 pKa = 11.84ASPHH81 pKa = 5.37PRR83 pKa = 11.84SRR85 pKa = 11.84RR86 pKa = 11.84KK87 pKa = 8.88WMHH90 pKa = 5.89HH91 pKa = 4.94
Molecular weight: 10.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2221 |
91 |
717 |
317.3 |
35.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.178 ± 0.527 | 2.296 ± 0.706 |
4.683 ± 0.444 | 6.529 ± 0.448 |
3.647 ± 0.368 | 5.583 ± 0.707 |
2.341 ± 0.662 | 3.917 ± 0.707 |
6.123 ± 1.178 | 11.571 ± 1.81 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.701 ± 0.248 | 3.332 ± 0.426 |
8.915 ± 1.103 | 4.728 ± 0.199 |
5.313 ± 0.872 | 7.564 ± 0.447 |
5.133 ± 0.644 | 5.853 ± 0.769 |
1.441 ± 0.35 | 3.152 ± 0.29 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
