
Methanonatronarchaeum thermophilum
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Methanonatronarchaeia; Methanonatronarchaeales; Methanonatronarchaeaceae; Methanonatronarchaeum
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
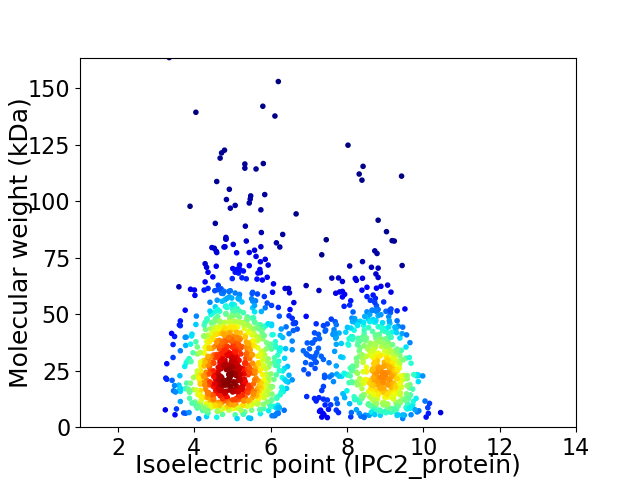
Virtual 2D-PAGE plot for 1510 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y3GAM7|A0A1Y3GAM7_9EURY Uncharacterized protein OS=Methanonatronarchaeum thermophilum OX=1927129 GN=AMET1_1228 PE=4 SV=1
MM1 pKa = 7.53VPRR4 pKa = 11.84KK5 pKa = 9.14YY6 pKa = 10.79LKK8 pKa = 10.81LLFVVCLLSSVLLVGGCVDD27 pKa = 3.44YY28 pKa = 11.32FEE30 pKa = 5.29SDD32 pKa = 3.06KK33 pKa = 11.28EE34 pKa = 4.1RR35 pKa = 11.84ITIVDD40 pKa = 3.55STGEE44 pKa = 4.17EE45 pKa = 3.84IEE47 pKa = 3.92INYY50 pKa = 9.21PVEE53 pKa = 4.08NVVTLTSDD61 pKa = 2.79SAEE64 pKa = 3.92AVRR67 pKa = 11.84ALGAEE72 pKa = 4.25DD73 pKa = 3.75KK74 pKa = 11.25VVGINDD80 pKa = 3.63YY81 pKa = 10.68MSGDD85 pKa = 3.19FWGDD89 pKa = 2.92LGEE92 pKa = 4.58IDD94 pKa = 4.17SVGNIFNPNPEE105 pKa = 4.64EE106 pKa = 4.07IASLEE111 pKa = 4.05PKK113 pKa = 9.74PDD115 pKa = 3.33LVLTYY120 pKa = 9.97TEE122 pKa = 4.16YY123 pKa = 11.14TEE125 pKa = 5.94DD126 pKa = 5.42LEE128 pKa = 7.12DD129 pKa = 4.0DD130 pKa = 4.66LKK132 pKa = 11.29PFGIDD137 pKa = 3.19VVRR140 pKa = 11.84LDD142 pKa = 4.04FYY144 pKa = 11.8KK145 pKa = 9.82MDD147 pKa = 4.74SIEE150 pKa = 4.15EE151 pKa = 4.22EE152 pKa = 4.06IEE154 pKa = 3.85ILGEE158 pKa = 3.9ILDD161 pKa = 4.53KK162 pKa = 11.1EE163 pKa = 4.4DD164 pKa = 3.7EE165 pKa = 4.4AEE167 pKa = 4.07EE168 pKa = 4.62LLNFYY173 pKa = 9.76NKK175 pKa = 10.26HH176 pKa = 4.77MDD178 pKa = 3.47EE179 pKa = 4.25VKK181 pKa = 10.69EE182 pKa = 3.7LAGDD186 pKa = 3.6INEE189 pKa = 4.5SEE191 pKa = 4.44VKK193 pKa = 10.23DD194 pKa = 3.74VYY196 pKa = 10.98IEE198 pKa = 4.57GFEE201 pKa = 4.12EE202 pKa = 4.49WSTASTNEE210 pKa = 4.08SNYY213 pKa = 8.79HH214 pKa = 4.55QVVEE218 pKa = 4.4LVGANNIAGDD228 pKa = 3.85RR229 pKa = 11.84DD230 pKa = 3.39RR231 pKa = 11.84TYY233 pKa = 9.86PTVSAEE239 pKa = 3.71WVLDD243 pKa = 3.94NNPDD247 pKa = 3.1AMMKK251 pKa = 9.82VVQDD255 pKa = 4.04SSVLGYY261 pKa = 10.51DD262 pKa = 3.71VNCTEE267 pKa = 5.28NAEE270 pKa = 4.04QMYY273 pKa = 10.91NDD275 pKa = 3.15IVDD278 pKa = 3.93RR279 pKa = 11.84EE280 pKa = 4.41GLSEE284 pKa = 3.97TDD286 pKa = 3.05AVKK289 pKa = 10.69NDD291 pKa = 3.35EE292 pKa = 4.58LVLVSQNVLSTMQNNIGTLIMAEE315 pKa = 3.93YY316 pKa = 9.86LYY318 pKa = 10.41PDD320 pKa = 3.87VYY322 pKa = 11.38EE323 pKa = 5.99DD324 pKa = 3.6IDD326 pKa = 3.56PMEE329 pKa = 3.86VHH331 pKa = 6.65EE332 pKa = 5.31EE333 pKa = 4.04YY334 pKa = 11.17LEE336 pKa = 4.52DD337 pKa = 3.64FHH339 pKa = 9.11GIEE342 pKa = 4.98YY343 pKa = 10.72DD344 pKa = 4.13GIWYY348 pKa = 9.64YY349 pKa = 11.67SKK351 pKa = 11.48
MM1 pKa = 7.53VPRR4 pKa = 11.84KK5 pKa = 9.14YY6 pKa = 10.79LKK8 pKa = 10.81LLFVVCLLSSVLLVGGCVDD27 pKa = 3.44YY28 pKa = 11.32FEE30 pKa = 5.29SDD32 pKa = 3.06KK33 pKa = 11.28EE34 pKa = 4.1RR35 pKa = 11.84ITIVDD40 pKa = 3.55STGEE44 pKa = 4.17EE45 pKa = 3.84IEE47 pKa = 3.92INYY50 pKa = 9.21PVEE53 pKa = 4.08NVVTLTSDD61 pKa = 2.79SAEE64 pKa = 3.92AVRR67 pKa = 11.84ALGAEE72 pKa = 4.25DD73 pKa = 3.75KK74 pKa = 11.25VVGINDD80 pKa = 3.63YY81 pKa = 10.68MSGDD85 pKa = 3.19FWGDD89 pKa = 2.92LGEE92 pKa = 4.58IDD94 pKa = 4.17SVGNIFNPNPEE105 pKa = 4.64EE106 pKa = 4.07IASLEE111 pKa = 4.05PKK113 pKa = 9.74PDD115 pKa = 3.33LVLTYY120 pKa = 9.97TEE122 pKa = 4.16YY123 pKa = 11.14TEE125 pKa = 5.94DD126 pKa = 5.42LEE128 pKa = 7.12DD129 pKa = 4.0DD130 pKa = 4.66LKK132 pKa = 11.29PFGIDD137 pKa = 3.19VVRR140 pKa = 11.84LDD142 pKa = 4.04FYY144 pKa = 11.8KK145 pKa = 9.82MDD147 pKa = 4.74SIEE150 pKa = 4.15EE151 pKa = 4.22EE152 pKa = 4.06IEE154 pKa = 3.85ILGEE158 pKa = 3.9ILDD161 pKa = 4.53KK162 pKa = 11.1EE163 pKa = 4.4DD164 pKa = 3.7EE165 pKa = 4.4AEE167 pKa = 4.07EE168 pKa = 4.62LLNFYY173 pKa = 9.76NKK175 pKa = 10.26HH176 pKa = 4.77MDD178 pKa = 3.47EE179 pKa = 4.25VKK181 pKa = 10.69EE182 pKa = 3.7LAGDD186 pKa = 3.6INEE189 pKa = 4.5SEE191 pKa = 4.44VKK193 pKa = 10.23DD194 pKa = 3.74VYY196 pKa = 10.98IEE198 pKa = 4.57GFEE201 pKa = 4.12EE202 pKa = 4.49WSTASTNEE210 pKa = 4.08SNYY213 pKa = 8.79HH214 pKa = 4.55QVVEE218 pKa = 4.4LVGANNIAGDD228 pKa = 3.85RR229 pKa = 11.84DD230 pKa = 3.39RR231 pKa = 11.84TYY233 pKa = 9.86PTVSAEE239 pKa = 3.71WVLDD243 pKa = 3.94NNPDD247 pKa = 3.1AMMKK251 pKa = 9.82VVQDD255 pKa = 4.04SSVLGYY261 pKa = 10.51DD262 pKa = 3.71VNCTEE267 pKa = 5.28NAEE270 pKa = 4.04QMYY273 pKa = 10.91NDD275 pKa = 3.15IVDD278 pKa = 3.93RR279 pKa = 11.84EE280 pKa = 4.41GLSEE284 pKa = 3.97TDD286 pKa = 3.05AVKK289 pKa = 10.69NDD291 pKa = 3.35EE292 pKa = 4.58LVLVSQNVLSTMQNNIGTLIMAEE315 pKa = 3.93YY316 pKa = 9.86LYY318 pKa = 10.41PDD320 pKa = 3.87VYY322 pKa = 11.38EE323 pKa = 5.99DD324 pKa = 3.6IDD326 pKa = 3.56PMEE329 pKa = 3.86VHH331 pKa = 6.65EE332 pKa = 5.31EE333 pKa = 4.04YY334 pKa = 11.17LEE336 pKa = 4.52DD337 pKa = 3.64FHH339 pKa = 9.11GIEE342 pKa = 4.98YY343 pKa = 10.72DD344 pKa = 4.13GIWYY348 pKa = 9.64YY349 pKa = 11.67SKK351 pKa = 11.48
Molecular weight: 39.83 kDa
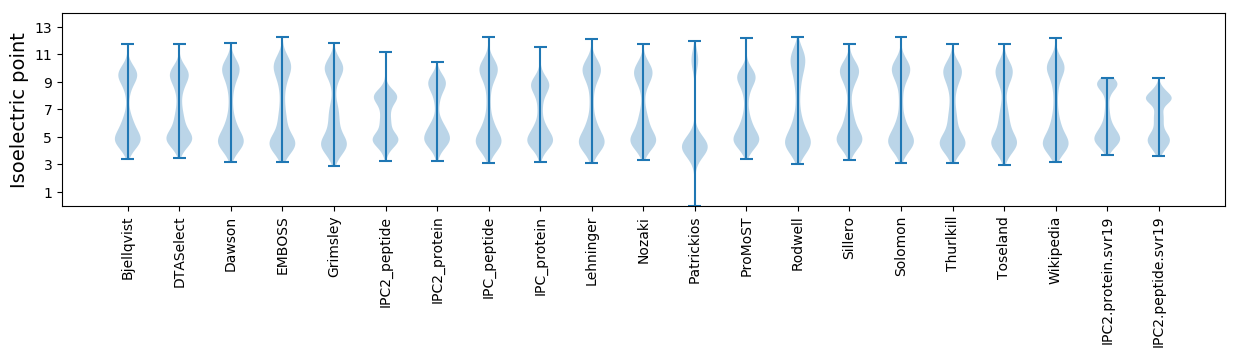
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y3GCC3|A0A1Y3GCC3_9EURY Uncharacterized protein OS=Methanonatronarchaeum thermophilum OX=1927129 GN=AMET1_0541 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.52SMYY5 pKa = 10.85SFIRR9 pKa = 11.84DD10 pKa = 2.9AWKK13 pKa = 10.3KK14 pKa = 9.11PRR16 pKa = 11.84EE17 pKa = 4.4GYY19 pKa = 8.44TRR21 pKa = 11.84EE22 pKa = 4.12LMWEE26 pKa = 4.27RR27 pKa = 11.84LQKK30 pKa = 9.42WRR32 pKa = 11.84SEE34 pKa = 4.11PTVKK38 pKa = 10.12RR39 pKa = 11.84VEE41 pKa = 3.99RR42 pKa = 11.84PTRR45 pKa = 11.84LDD47 pKa = 3.07KK48 pKa = 11.08ARR50 pKa = 11.84KK51 pKa = 8.7NGYY54 pKa = 8.07KK55 pKa = 10.16AKK57 pKa = 10.27EE58 pKa = 4.09GFVVARR64 pKa = 11.84ARR66 pKa = 11.84VRR68 pKa = 11.84RR69 pKa = 11.84GTRR72 pKa = 11.84RR73 pKa = 11.84KK74 pKa = 10.12SRR76 pKa = 11.84FKK78 pKa = 10.7KK79 pKa = 9.56GRR81 pKa = 11.84KK82 pKa = 6.64PSKK85 pKa = 9.8MGLEE89 pKa = 4.48KK90 pKa = 9.74ITPGKK95 pKa = 9.49SLQRR99 pKa = 11.84IAEE102 pKa = 4.06EE103 pKa = 3.76RR104 pKa = 11.84TSRR107 pKa = 11.84KK108 pKa = 9.25FPNLRR113 pKa = 11.84VLASYY118 pKa = 9.37WVGEE122 pKa = 4.43DD123 pKa = 5.13GRR125 pKa = 11.84QKK127 pKa = 8.93WYY129 pKa = 10.38EE130 pKa = 3.95VIMVDD135 pKa = 3.39THH137 pKa = 6.97HH138 pKa = 7.22PAIKK142 pKa = 10.2NDD144 pKa = 3.37SDD146 pKa = 4.16INWICSDD153 pKa = 2.97KK154 pKa = 11.04HH155 pKa = 6.16KK156 pKa = 11.13NRR158 pKa = 11.84AFRR161 pKa = 11.84GLTTVGKK168 pKa = 9.9DD169 pKa = 3.25GRR171 pKa = 11.84GLSEE175 pKa = 3.97RR176 pKa = 11.84GKK178 pKa = 9.09GTEE181 pKa = 3.71KK182 pKa = 9.85TRR184 pKa = 11.84PSIRR188 pKa = 11.84SNEE191 pKa = 4.24GKK193 pKa = 10.77GKK195 pKa = 10.49
MM1 pKa = 7.44KK2 pKa = 10.52SMYY5 pKa = 10.85SFIRR9 pKa = 11.84DD10 pKa = 2.9AWKK13 pKa = 10.3KK14 pKa = 9.11PRR16 pKa = 11.84EE17 pKa = 4.4GYY19 pKa = 8.44TRR21 pKa = 11.84EE22 pKa = 4.12LMWEE26 pKa = 4.27RR27 pKa = 11.84LQKK30 pKa = 9.42WRR32 pKa = 11.84SEE34 pKa = 4.11PTVKK38 pKa = 10.12RR39 pKa = 11.84VEE41 pKa = 3.99RR42 pKa = 11.84PTRR45 pKa = 11.84LDD47 pKa = 3.07KK48 pKa = 11.08ARR50 pKa = 11.84KK51 pKa = 8.7NGYY54 pKa = 8.07KK55 pKa = 10.16AKK57 pKa = 10.27EE58 pKa = 4.09GFVVARR64 pKa = 11.84ARR66 pKa = 11.84VRR68 pKa = 11.84RR69 pKa = 11.84GTRR72 pKa = 11.84RR73 pKa = 11.84KK74 pKa = 10.12SRR76 pKa = 11.84FKK78 pKa = 10.7KK79 pKa = 9.56GRR81 pKa = 11.84KK82 pKa = 6.64PSKK85 pKa = 9.8MGLEE89 pKa = 4.48KK90 pKa = 9.74ITPGKK95 pKa = 9.49SLQRR99 pKa = 11.84IAEE102 pKa = 4.06EE103 pKa = 3.76RR104 pKa = 11.84TSRR107 pKa = 11.84KK108 pKa = 9.25FPNLRR113 pKa = 11.84VLASYY118 pKa = 9.37WVGEE122 pKa = 4.43DD123 pKa = 5.13GRR125 pKa = 11.84QKK127 pKa = 8.93WYY129 pKa = 10.38EE130 pKa = 3.95VIMVDD135 pKa = 3.39THH137 pKa = 6.97HH138 pKa = 7.22PAIKK142 pKa = 10.2NDD144 pKa = 3.37SDD146 pKa = 4.16INWICSDD153 pKa = 2.97KK154 pKa = 11.04HH155 pKa = 6.16KK156 pKa = 11.13NRR158 pKa = 11.84AFRR161 pKa = 11.84GLTTVGKK168 pKa = 9.9DD169 pKa = 3.25GRR171 pKa = 11.84GLSEE175 pKa = 3.97RR176 pKa = 11.84GKK178 pKa = 9.09GTEE181 pKa = 3.71KK182 pKa = 9.85TRR184 pKa = 11.84PSIRR188 pKa = 11.84SNEE191 pKa = 4.24GKK193 pKa = 10.77GKK195 pKa = 10.49
Molecular weight: 22.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
425233 |
31 |
1462 |
281.6 |
31.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.542 ± 0.064 | 0.987 ± 0.027 |
5.926 ± 0.065 | 8.586 ± 0.086 |
3.497 ± 0.051 | 7.076 ± 0.061 |
1.816 ± 0.03 | 8.741 ± 0.068 |
8.675 ± 0.1 | 8.696 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.233 ± 0.028 | 4.904 ± 0.053 |
3.851 ± 0.039 | 3.203 ± 0.047 |
4.193 ± 0.051 | 5.451 ± 0.051 |
5.809 ± 0.06 | 6.694 ± 0.079 |
0.875 ± 0.023 | 3.245 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
